Figures & data
Figure 1. Geometric mean frequencies (GMFs) of (A) gE-specific CD4+ T cells and (B) ratios of GMFs from different adjuvanted-vaccine groups. Spleens (Experiment 1, N=8 and Experiment 2, N = 11; spleens pooled from 2 mice) were sampled at 30 d after the second vaccine dose (30dPII). The frequency of gE-specific CD4+ T cells was calculated as a percentage of cytokine-positive CD4+ T cells divided by all CD4+ T cells. Error bars represent 95% confidence intervals. In Experiments 1 and 2, the frequencies of antigen-specific CD4+ T cells in the NaCl group were either close to or below the cut-off for the assay (GMFs were 0.3% and 0.05%, respectively). In (B), horizontal gray reference line indicates a ratio = 1, and asterisks indicate significant differences from 1 (** p<0.01; *** p<0.001). Antigen-specific T cells were evaluated in splenocyte-restimulation cultures as described previously,Citation13 but with some modifications. Briefly, splenocyte cultures (1106 cells per well of 96-well plate) were prepared from spleens of 2 mice and were incubated for 2 hours in the presence of gE peptides spanning the complete gE protein (6315-mer peptides, 11 amino-acid overlap) and then incubated ˜18 hours in the presence of brefeldin A. Subsequently, the cells were stained with fluorescent-monoclonal antibodies specific for CD4 and after permeabilization, for intracellular-cytokines IL-2 and IFN-γ. All antibodies were obtained from BD Biosciences, Belgium. Flow cytometry was performed using LSR II Facs (BD Biosciences, Belgium) and analyzed using FlowJo software (FlowJo, LLC, OR, USA). Statistical calculations were based on an analysis of variance with 2 factors (vaccine group, experiment) on log10 values using a heterogeneous variance model (i.e., identical variances were not assumed for the different levels of the factor). Estimates of the geometric-mean ratios between groups and their 95% confidence intervals (CI) were obtained using back-transformation of log10 values. Adjustments for multiple testing were performed using Tukey's method. All analyses were performed using SAS software (Version 9.2, SAS Institute Inc., NC, USA).
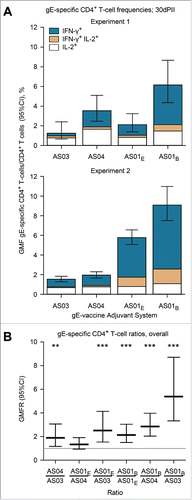
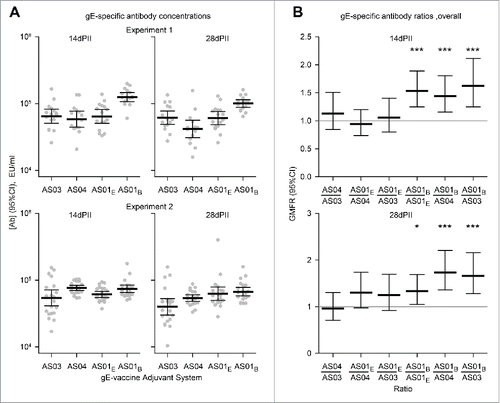
Figure 2. Geometric mean concentrations (GMCs) of (A) gE-specific antibodies and (B) ratios of GMCs (GMFRs) from different adjuvanted-vaccine groups. Sera (Experiment 1, N = 16 1 and Experiment 2, N = 22) were sampled at 14 and 28 d after the second vaccine dose (14dPII and 28dPII, respectively). Error bars represent 95% confidence intervals. Antigen-specific antibodies were not detected in the NaCl group (concentrations were below the cut-off of the assay; i.e.<500 EU/ml). In (B), horizontal gray reference lines indicate a ratio = 1, and asterisks indicate significant differences from 1 (*p<0.05; *** p<0.001). Antigen-specific antibody concentrations (in EU/ml and defined by internal standards) were measured by ELISA as previously described.Citation13 Statistical calculations were performed as described in .