Figures & data
Figure 1. Serum bactericidal assay with human complement. Figure adapted from Ghandi, et al. Postgraduate Medicine, 2016.Citation82
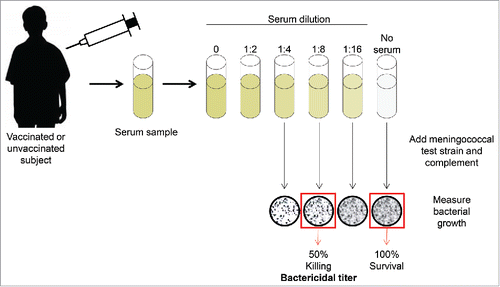
Table 1. MenB test strains used for the assessment of bivalent rLP2086.
Figure 2. Phylogenetic relationship of different fHBP subgroups in the extended MenB SBA strain pool. Representation of the phylogeny of fHBP is based on a clustalW alignment and drawn with MEGA 4.2. The relative phylogenetic position of subfamily A and B fHBP variants, as well as the 6 major subgroups of fHBP, are outlined. The phylogenetic position of fHBP variants expressed by the 4 primary and 10 secondary SBA test strains, as well as 2 vaccine variants, is highlighted. The numbers beneath each of the fHBP subgroups represent the number of isolates, number of unique fHBP variants, and average amino acid percent identity from the respective subgroups. The scale bar represents phylogenetic distance based on the deduced fHBP protein sequence. Phylogenetic relationship of MenB fHBP variants first described by Murphy, et al. J Infect Dis. 2009.Citation30
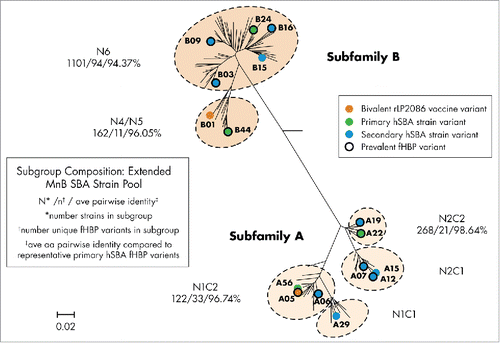
Table 2. Bactericidal activity of bivalent rLP2086 immune sera against university outbreak strains in the United States.Footnote*
Figure 3. Activity of rLP2086 immune sera against fHBP B133 isolates in exploratory hSBAs. Isolates tested were from the University E outbreak (PMB5543 and PMB5544) and from the Netherlands (PMB5507). Sera are from 15 subjects vaccinated with bivalent rLP2086. Sera were collected before vaccination, after the first vaccination (PD1), after the second vaccination (PD2), and after the third vaccination (PD3). Responder rates based on individual titers ≥1:4 are indicated. Open rectangles represent geometric mean titers with 95% CIs (error bars). fHBP = factor H binding protein; hSBA = serum bactericidal assay using human complement; PD = postdose.
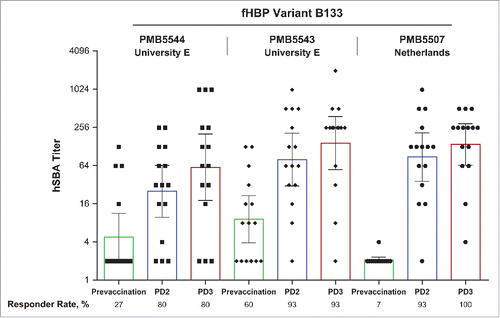
Figure 4. MATS assay. Bacteria are grown overnight. A bacterial suspension is grown to a specific OD and detergent is added to the suspension to extract the capsule and expose the antigens. Serial dilutions of the extract are tested by ELISA. fHBP, NHBA, and NadA coverage is assessed by defining a relative potency of the tested strain versus a reference strain for each antigen and then extrapolating a relative potency to a positive bactericidal threshold. PCR genotyping is used to identify PorA. ELISA = enzyme-linked immunosorbent assay; fHBP = factor H binding protein; MATS = Meningococcal Antigen Typing System; NadA = Neisserial adhesin A; NHBA = Neisserial heparin binding antigen; OD = optical density; PCR = polymerase chain reaction; PorA = porin A.
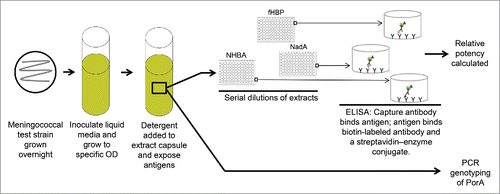
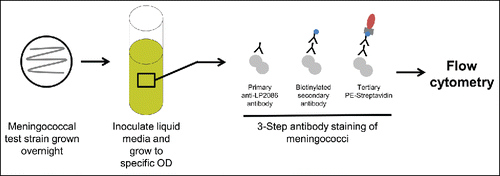
Figure 5. MEASURE assay. Bacteria are grown overnight using standard hSBA procedures. Bacteria are then stained using a 3-step antibody staining method. Flow cytometry is used to selectively quantify the concentration of surface-expressed fHBP. fHBP = factor H binding protein; hSBA = serum bactericidal assay using human complement; MEASURE = Meningococcal Antigen Surface Expression; OD = optical density.