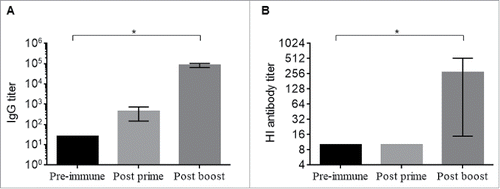
Figures & data
Figure 1. Immunoblot profiles of (A) HA-MYnative, (B) HA-MY and (C) HA1-MY in extracts of infiltrated N. benthamiana leaves at 8 dpi, using an anti-4xHis antibody. M: Molecular weight marker, Lane 1: HA Indonesia 15 ng, Lane 2: HA Indonesia 60 ng, Lane 3: PTP, Lane 4: TSP, Lane 5: TST.
Figure 2. Three-column purification process analyzed by SDS-PAGE and Western blot. (A) Coomassie-stained SDS-PAGE gel, (B) Western blot analysis using an anti-4xHis antibody, and (C) Western blot analysis using an anti-RuBisCO antibody. M: Molecular weight marker, Lane 1: E fraction from IMAC, Lane 2: FT fraction from Butyl column, Lane 3: FT fraction from Q column, Lane 4: E fraction from Q column, Lane 5: TSP.
Figure 3. Two-column purification process analyzed by SDS-PAGE and Western blot. (A) Coomassie-stained SDS-PAGE gel, (B) Western blot analysis using an anti-4xHis antibody, and (C) Western blot analysis using an anti-RuBisCO antibody. M: Molecular weight marker, Lane 1: E fraction from IMAC, Lane 2: FT fraction from Q column, Lane 3: E fraction from Q column.
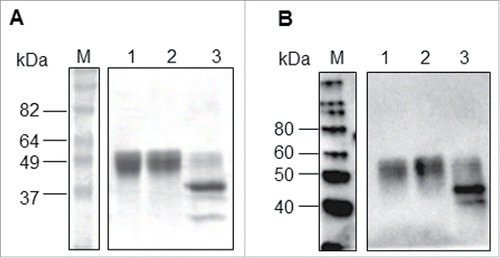
Figure 4. Deglycosylation of HA1-MY treated with PNGase F. (A) Coomassie-stained SDS-PAGE gel and (B) Western blot analysis using an anti-A/Vietnam/1194/04 antibody. M: Molecular weight marker, Lane 1: Non-treated HA1-MY, Lane 2: Mock-treated HA1-MY without PNGase F, Lane 3: PNGase F-treated HA1-MY.