Figures & data
Figure 1. Magnitude of human immunodeficiency virus (HIV)-1 subtype B Nef core specific T cell responses in 29 HIV-1B-infected individuals. Measured as interferon (IFN)-γ enzyme-linked immunospot (ELISPOT), represented as spot-forming units (SFU)/106 peripheral blood mononuclear cells.
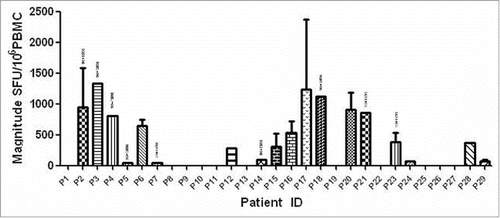
Figure 2. Magnitude and frequency of 10 HIV-1 subtype B Nef core OLPs. The total magnitude of T cell responses specific for each tested peptide is shown as bars in the left part of the graph. The frequency of each tested peptide is shown as bars on the right part of the graph.
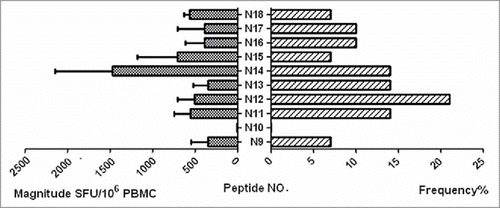
Figure 3. The heatmap for all patients and all peptides provide full picture with the ELISPOT responses. The right ordinate is the patient NO., The bottom abscissa is the peptide NO. The heatmap should define the cut-off as the detection limit.
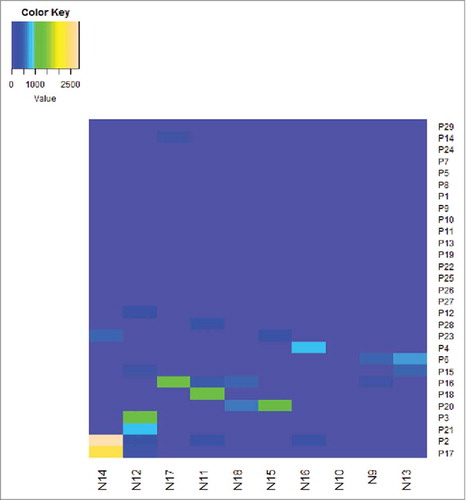
Figure 4. ELISPOT magnitude is associated with CD4+T cell count and viral load. The associations between ELISPOT magnitude (total spot forming units per 1 Million PBMCs and CD4+T cell count or viral load) were analyzed by Spearman's correlation. The Nef core-specific response was compared with CD4+T cell count (a) and viral load (b). P value is corrected by FDR.
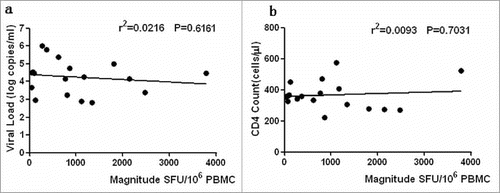
Figure 5. ELISPOT breadth is associated with CD4+T cell countand viral load. The associations between ELISPOT breadth [the number of reacting OLPs] and CD4+T cellcount (a) or viral load (b) were analyzed using the Kruskal-Wallis test. (n) Refers to the number of individuals. P value is corrected by FDR.
![Figure 5. ELISPOT breadth is associated with CD4+T cell countand viral load. The associations between ELISPOT breadth [the number of reacting OLPs] and CD4+T cellcount (a) or viral load (b) were analyzed using the Kruskal-Wallis test. (n) Refers to the number of individuals. P value is corrected by FDR.](/cms/asset/a72f38c1-c3dc-480d-811a-e5150fc3d58f/khvi_a_1340138_f0005_b.gif)
Figure 6. Comparison of CD4 count, VL, Magnitude and Breadth between DQB1*06 positive and negative group. P value is corrected by FDR.
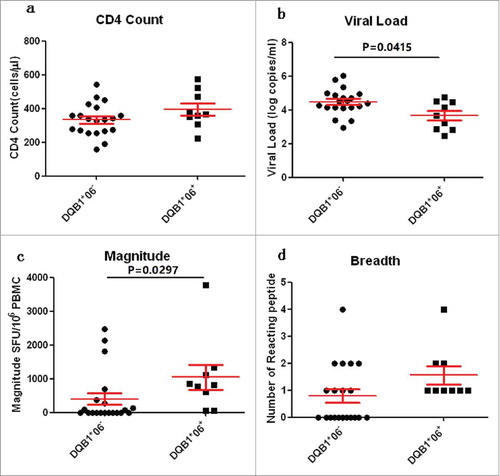
Figure 7. DQB1*06 allele-specific enzyme-linked immunospot (ELISPOT) magnitude were compared with viral load (VL) and CD4+ T cell count. P value is corrected by FDR.
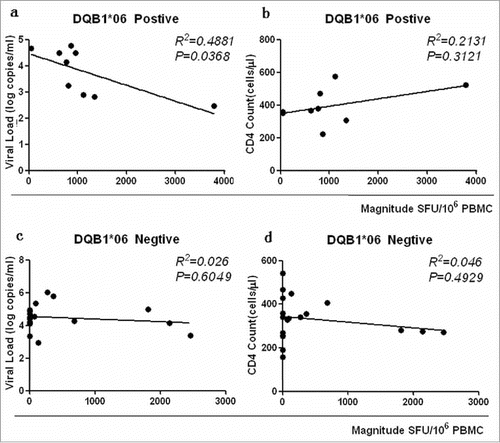
Table 1. Characteristics of the study patient cohort.
Table 2. Nef Core OLPs sequence, magnitude and breath.
