Figures & data
Figure 1. Differential genomic signatures in PBMCs between the non-responder and responder groups at 5 time points. (A-E) PBMCs from 3 participants of each group was conducted tanscriptome analysis at 5 time points, i.e. pre-vaccination, 3rd, 7th, 28th day post the first dose vaccination and 7th day post the second dose vaccination. DEGs exhibited significant differences (Fold Change ≥2.0 and p-value <0.05) between the two groups at all time points. Transcript expressions were transformed to z-score and depicted in green to black to red color scale.
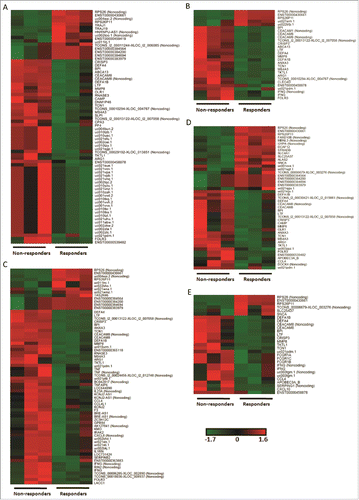
Figure 2. Interaction networks of proteins derived from differentially expressed coding RNAs. STRING database (http://string-db.org/) was used to assess the integration of protein-protein interactions. All DEGs discovered from each time point were analyzed for the interaction of encoded proteins. The numbers of lines represented the closeness of these proteins. Line thickness indicates the strength of data support from the database.
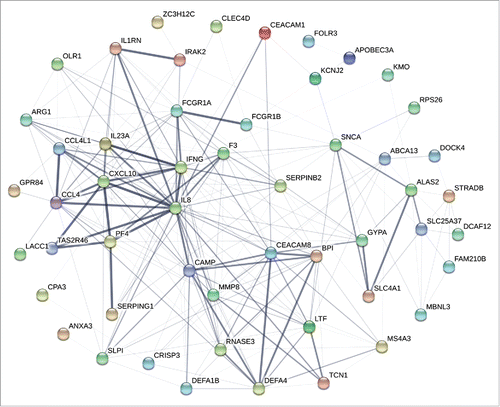
Figure 3. Comprehensive analyses and dynamic changes of the DEGs identified in non-responder and responder groups at 5 different time points. (A-B) Venn diagrams indicated the numbers of persistent up- or down-regulated expressed genes in non-responders at all time points, compared with responders. (C) Heat map showed the fold change of 11 up-regulated and 3 down-regulated DEGs in non-responders with persistent significant expression at all 5 time points, compared with responders. The change threshold was set to 2-fold.
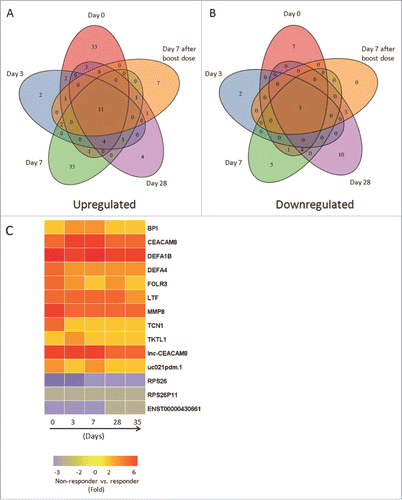
Figure 4. Validation of up-regulated expressed genes by qRT-PCR. The expression of 9 up-regulated genes (Fold Change ≥2.0) identified by microarray assay (in black color) at 5 time points was re-evaluated by qRT-PCR (in gray color). Eight genes in qRT-PCR results confirmed the data of the microarray.
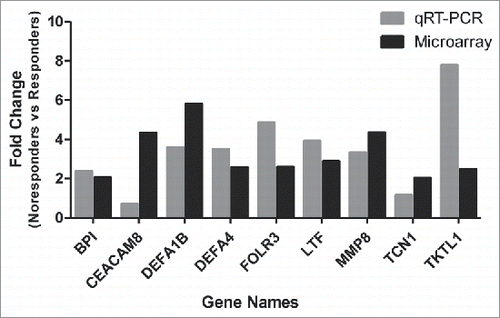
Figure 5. GO analysis for differentially expressed genes between non-responders and responders at 5 different time points. (A-E)The top 10 biological process of DEGs at the time point of pre-vaccination, 3rd, 7th, 28th day post the first dose vaccination and 7th day post the second dose vaccination, respectively.
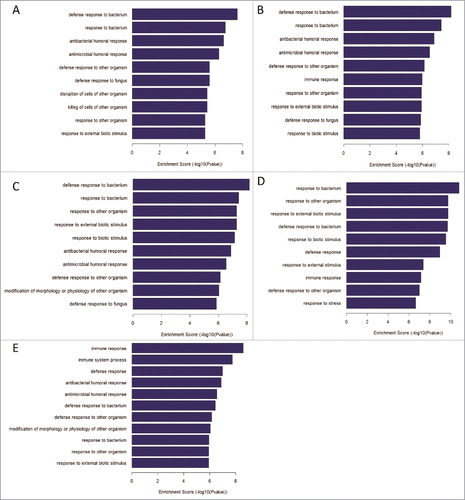
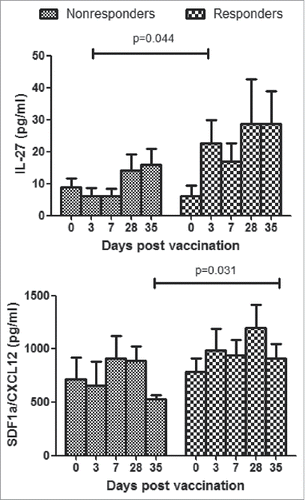
Figure 6. Cytokine responses in plasma of HBV vaccine non-responders and responders. Plasmas from all participants were assayed by Luminex. Concentrations of cytokines were expressed as means ± SEM. Statistical analysis was performed using Mann Whitney test for non-responder and responder group at each time point.