Figures & data
Figure 1. Crystal structure of the complex between meningococcal FHbp and an FH fragment comprising domains 6 and 7. The two molecules were separated along the Y-axis to show the respective binding surfaces (color). Mutations to decrease FH binding to FHbp target the top, green surface of FHbp. Coordinates are from PDB ID 2w80.Citation38 Figure was constructed with PyMol (The PyMOL Molecular Graphics System, Version 2.3 Schrödinger, LLC)
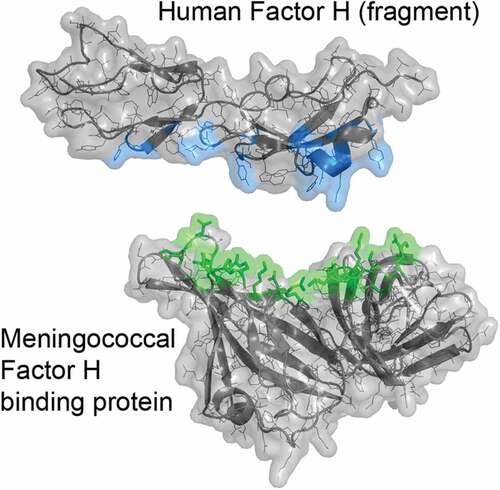
Figure 2. Relationship between decreases in FH binding of mutant FHbp antigens and serum bactericidal antibody responses, each relative to the respective wild-type FHbp antigen. Magenta circles represent sub-family A (ID22) antigens and blue squares depict sub-family B (ID1) antigens. Fold changes in FH binding were measured by surface plasmon resonance, with the exception of K219N and G220S, which were measured by ELISA. The data from the latter two mutants are from proteins with two additional stabilizing substitutions as previously described.Citation52,Citation61 It should be noted that the increases in bactericidal activity elicited by mutants as compared to the wild-type antigens depend on the FHbp sequence and expression in the target strain.Citation43,Citation58.
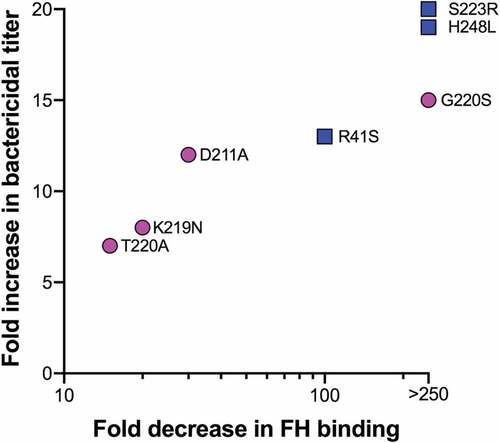
Figure 3. FH inhibition by macaque serum antibodies elicited by a native OMV vaccine with over-expressed mutant FHbp. After two vaccine doses, antibodies from all 13 macaques immunized with the native OMV vaccine inhibited FH binding to purified, recombinant FHbp, whereas antibodies from animals to a licensed control vaccine did not inhibit and some enhanced FH binding. The p-value for a paired t-test is shown. Macaques immunized with aluminum hydroxide (Al(OH)3) as a negative control did not show significant inhibition or enhancement. Figure adapted from ref.Citation56
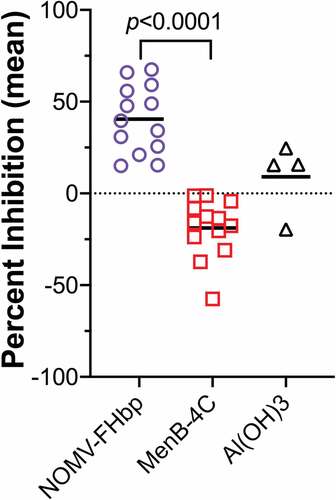
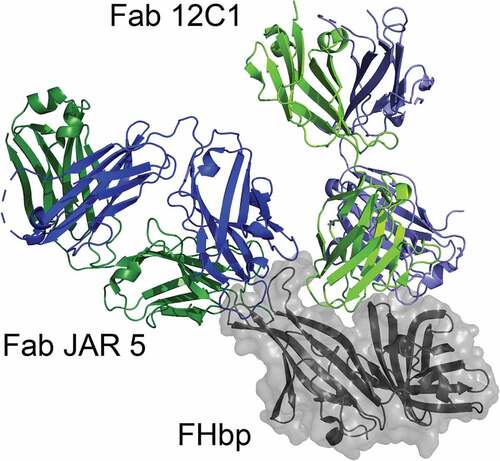
Figure 4. Molecular model of two antibody Fab fragments bound to FHbp. The figure was generated by superimposing the FHbp molecules from two experimentally determined structures of FHbp-Fab complexes (Fab 12C1; PDB ID 2YPV and Fab JAR 5; 5T5F).Citation73,Citation75 FHbp is shown in gray with surface rendering and the heavy and light chains of each Fab are shown in blue and green, respectively. JAR 5 binds to a region of the amino-terminal domain of FHbp (top left) and 12C1 binds to a surface that overlaps the FH binding site (top). The Fabs bind to non-overlapping epitopes and the respective MAbs elicit cooperative bactericidal activity.Citation75 Figure was constructed with PyMol (The PyMOL Molecular Graphics System, Version 2.3 Schrödinger, LLC)