Figures & data
Figure 1. Diagrammatic representation of inductive and effector sites for mucosal immunity. Two most important mucosal inductive sites are gut-associated lymphoid tissue (GALT) and nasopharynx-associated lymphoid tissue (NALT). These inductive sites are lined with follicle-associated epithelium which consists of microfold (M) cells responsible for the transport of antigens to the antigen presenting cells (APCs). These APC’s then trigger the cellular immunity by activating effector T cells which in turn elicit the IgA class switching in follicular plasma B cells. These IgA producing B cells then reach the effector sites through systemic circulation and release secretory IgA (sIgA). The polymeric immunoglobulin receptor (pIgR) located at the basal surface of effector sites, such as lamina propria etc. transfers sIgA to the luminal surface, where it inhibits the pathogen by three different mechanisms, namely, immune exclusion, antigen excretion, and intracellular neutralization
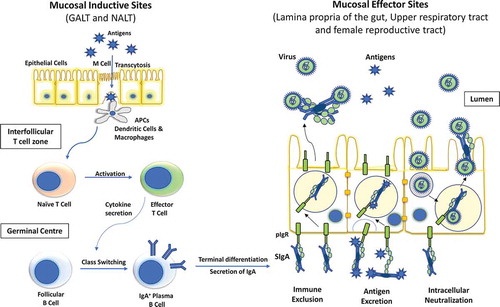
Table 1. SARS-CoV and MERS-CoV mucosal vaccines based on different vaccine platforms*
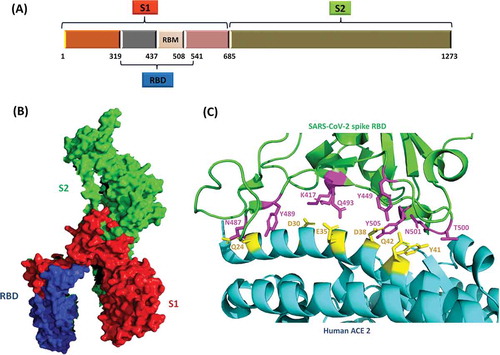
Figure 2. (a) Schematic of SARS-CoV-2 S protein and its subunits. S protein is the major determinant of receptor binding and pathogenesis in SARS-CoV-2. (b) SARS-CoV-2 S protein monomer (PDB ID: 6VXX). The S1 domain which contains the receptor binding domain (RBD) and receptor binding motif (RBM), is critical to the viral infectivity as it initiates the attachment of viral particle to the host cell. The S2 domain is responsible for the fusion of viral and host membranes leading to internalization of the virus. (c) Receptor binding domain (RBD) of SARS-CoV-2 bound to its host cell receptor, human angiotensin-converting enzyme 2 (hACE2) via specific amino-acid interactions (PDB ID: 6M0J). Interacting residues are shown as sticks at RBD-hACE2 interface.Citation70