Figures & data
Figure 1. Nanoparticles containing autoantigen (HEL46-61) and rapamycin induced tolerogenic DCs. (a) PEG-PLA nanoparticles containing autoantigen (HEL46-51) and rapamycin were constructed via double emulsion method. (b) Size distribution of NPHEL46-51/Rapa. (c) Uptake of NPHEL46-51/Rapa by mouse bone marrow dendritic cells (BMDCs). HEL46-51 was labeled with Cy5. (d) Colocalization of Cy5-HEL46-51 peptide (red) and MHC II molecules (green) in BMDCs at 24 h after the transfection of the cells with NPHEL46-51/Rapa. The percentages of CD80+ (e) and CD86+ (f) BMDCs post different treatments. The mean fluorescence intensity (MFI) of CD80 (g) and CD86 (h) on the surface of NPHEL46-51/Rapa-treated BMDCs. The absolute number of CD80+ (i) and CD86+ (j) BMDCs after different treatments. These experiments were repeated three times independently with similar results. Data were presented as mean ± SD and the statistical difference was calculated using one-way ANOVA with a Tukey’s post hoc test **p < .01, ***p < .001
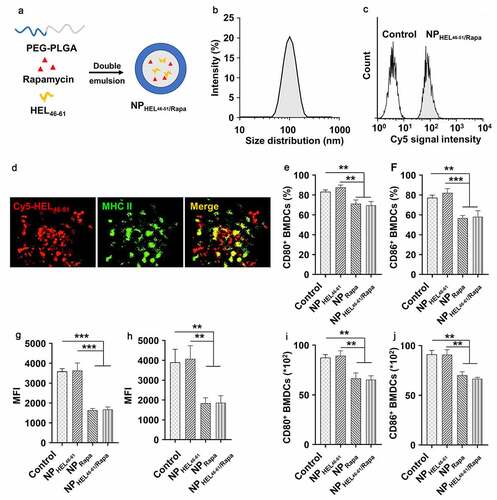
Figure 2. NPHEL46-61/Rapa -treated BMDCs suppressed antigen-specific CD4+ T cell proliferation and induced Treg differentiation. (a) Schematic illustration showing how the ability of NPHEL46-51/Rapa-treated DCs to induce the proliferation and differentiation of CD4+ T cells. (b) The proliferation of CD4+ T cells after coculture with treated BMDCs was examined by CFSE (Carboxyfluorescein diacetate succinimidylester) T cell proliferation assay. Flow cytometric analysis (c) and statistical data obtained in three replicate experiments in which the percentages of Treg cells (CD4+Foxp3+) in CD4+ T cells (d) and the absolute numbers and the percentage of Foxp3+ CD4 T cells (e) after coculture were determined. These experiments were repeated three times independently with similar results and the statistical difference was calculated using one-way ANOVA with a Tukey’s post hoc test. Data were presented as mean ± SD. n = 3, *p < .05, **p < .01, ***p < .001. ns, no significant difference
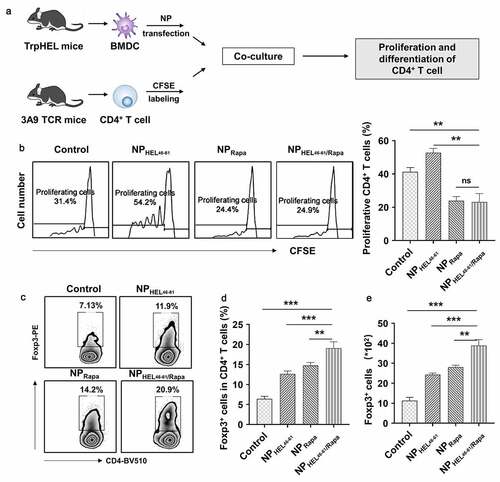
Figure 3. NPHEL46-61/Rapa ameliorated vitiligo and promoted Treg in TCR-TrpHEL mice. (a) Therapeutic scheme. TrpTEL mice were treated with NPHEL46-61/Rapa or other formulations at age of four weeks. Mice were treated three times per week for 4 weeks, and the vitiligo scores were recorded. (b) Weekly vitiligo scores of transgenic TCR-TrpHEL mice. (c) The representative staining of destroyed melanocytes for each type of treatment. Representative images of mice with vitiligo post treatment. Active caspase-3 (red; cytoplasmic) as an apoptosis indicator. Scale bar, 50 μm. Percentages of CD4+CD25+FOXP3+ Treg in CD4+ T cells in lymph (d), spleen (e) and blood (f) and skin (g) at 10th week. Data represent means ± SD. Statistical difference was calculated using one-way ANOVA with a Tukey’s post hoc test for the data in C-G, and two-way ANOVA with a Dunnett’s post hoc test for the data in B. n = 6, **p < .01, ***p < .001. ns, no significant difference
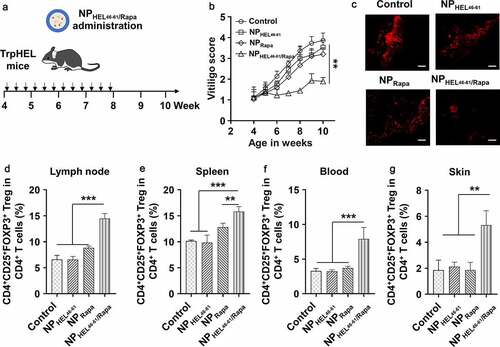
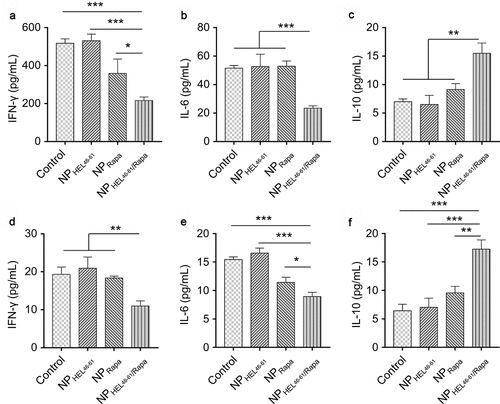
Figure 4. NPHEL46-61/Rapa enhanced IL-10 expression and suppressed IFN-γ and IL-6 expression in TCR-TrpHEL mice. (a–c) ELISA analysis of the production of IFN-γ (a), IL-6 (b) and IL-10 (c) in lymph nodes of TrpTEL mice treated with different formulations. (d–f) ELISA analysis of the production of IFN-γ (d), IL-6 (e) and IL-10 (f) in the spleens of TrpTEL mice treated with different formulations. Data represent means ± SD and the statistical difference was calculated using one-way ANOVA) with a Tukey’s post hoc test. n = 6, *p < .05, **p < .01, ***p < .001. ns, no significant difference