Figures & data
Figure 1. Distribution by age groups of all the tested MenB isolates for the two epidemiological years 2013–2014 and 2018–2019. The numbers of isolates per age group and per year are shown. Due to multiple comparisons, significance level was 0.007.
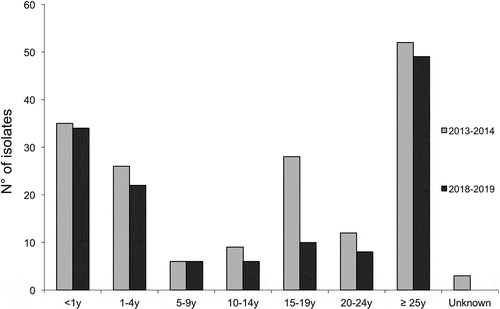
Figure 2. Distribution of all the tested isolates according to their clonal complexes (CC). The numbers of isolates per CC and per year are shown. UA, unassigned isolates to any of the currently defined CC.
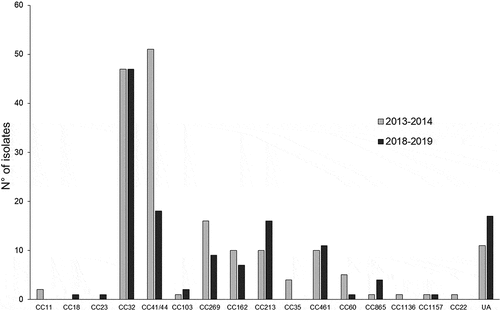
Table 1. Peptide identification number according to PubMLST
Figure 3. Distribution (in number of isolates) of fHbp (a) and NHBA (b) peptides of all the tested MenB isolates for the two epidemiological years 2013–2014 and 2018–2019.
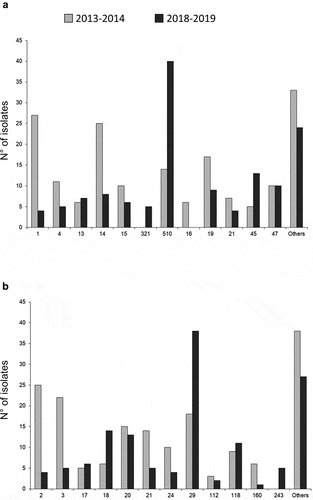
Table 2. Coverage rates prediction for the tested isolates according to MATS (with 95% CI) and gMATS (LL-UL)
Figure 4. gMATS-based coverage distribution of isolates by clonal complexes represented by more than 5 isolates in each period (two bars per CC; on the left the year 2013–2014 and on the right the year 2018–2019). The isolates of each CC were categorized in covered, unpredictable or non-covered as indicated. The coverage rate with the LL and UL for each CC are indicated above each corresponding bar.
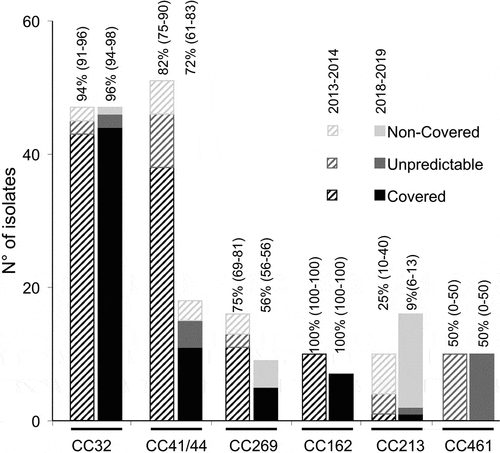
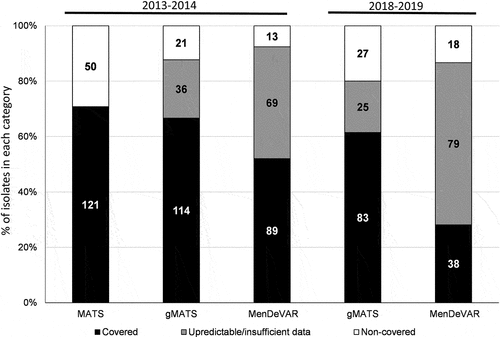
Figure 5. Prediction of 4CMenB coverage by MATS, gMATS and MenDeVAR (for the epidemiological year 2013–2014) and by gMATS and MenDeVAR (for the epidemiological year 2018–2019) of all the tested MenB isolates. The percentages of isolates within each predicted category are shown. For MenDeVAR, covered isolates accounted for both ”exact-match” + ”cross-reactive”. The number of isolates is indicated in the corresponding bars. The estimations of MATS and gMATS coverage are depicted in the .