Figures & data
Figure 1. Spike-specific serum IgG responses following single dose intranasal vaccination with AdCOVID. K18-hACE2 mice (8 mice/group) were intranasally vaccinated with 2.2E + 09 ifu AdCOVID or vehicle control on day 0. All mice were challenged on day 32 with 2.0E + 04 PFU of SARS-CoV-2 strain USA/WA-1/2020 by the intranasal route. (a) Sera were collected on day 21 and analyzed individually for quantification of spike-specific IgG by CBA. (b) Body weight was measured daily. Four days post-challenge (DPI +4), spike-specific IgG in the (c) serum and (d) nasal wash was quantified by CBA. IgG results are expressed in either µg/ml or ng/ml as indicated. Bars represent mean ± SEM. Body weights are presented as the mean ± SEM. Lowest limit of quantification (LLOQ). Statistical analyses were performed with unpaired Student’s t tests. ***, P < .001; ****, P < .0001.
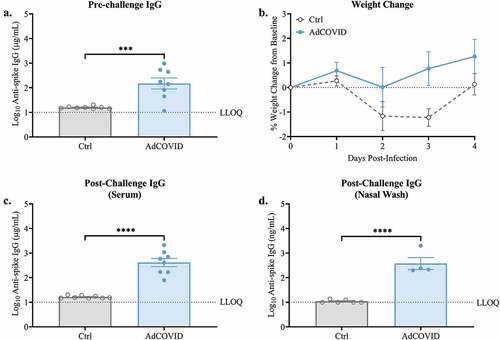
Figure 2. AdCOVID limits viral load in the nasal cavity following intranasal SARS-CoV-2 challenge. K18-hACE2 mice were intranasally vaccinated with 2.2E + 09 ifu AdCOVID or vehicle control on day 0, challenged on day 32 with (a-d) 2.0E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020 and analyzed between 0–4 days post-infection or (E-H) challenged on day 32 with 0.5E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020 and analyzed on DPI +3. (a-d) Nasal swipes (n = 8 mice/group) were taken daily for qRT-PCR quantification of (A) total viral RNA and (b) subgenomic N RNA. On DPI +4, animals (n = 4–6 mice/group) were euthanized, and nasal wash was collected for qRT-PCR quantification of (c) total viral N RNA and (d) subgenomic N RNA. (e-h) Spike-specific IgG antibodies measured in serum (panel E, n = 11 mice/group) on day 21 post-vaccination. Spike-specific IgA antibodies measured in BAL and nasal wash on DPI +3 (panel F, n = 4–5 mice/group). qRT-PCR quantification of (g) total viral N RNA and (h) subgenomic N RNA in nasal wash on DPI +3 (n = 4–5 mice/group). Viral quantitation presented as RNA copies. Bars represent mean ± SEM. Lowest limit of quantification (LLOQ). Statistical analyses were performed with unpaired Student’s t tests. **, P < .01; ***, P < .001; ****, P < .0001.
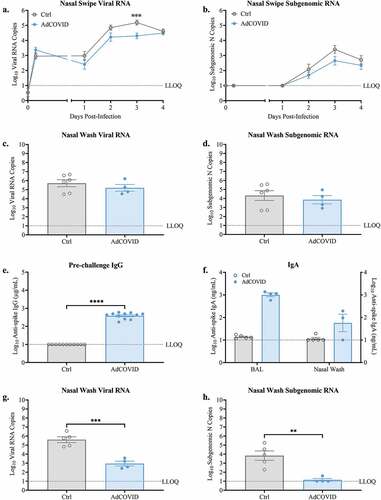
Figure 3. AdCOVID limits viral replication in the lungs following intranasal SARS-CoV-2 challenge. AdCOVID-vaccinated and control K18-hACE2 mice were challenged on day 32 with either (a, b) 2.0E + 04 PFU or (c, d) 0.5E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020. On (a-b) DPI +4 or (c, d) DPI +3, animals were euthanized, and both lung lobes were harvested. (A, C) Total viral N RNA was quantified by qRT-PCR. Results are expressed as RNA copies. (b, d) Infectious virus was quantified in the lungs by plaque assay. Bars represent mean ± SEM. Lowest limit of quantification (LLOQ). Statistical analyses were performed with unpaired Student’s t tests. *, P < .05; **, P < .01; ***, P < .001; ****, P < .0001.
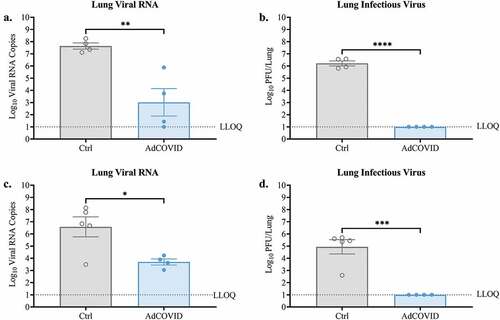
Figure 4. AdCOVID protects mice from SARS-CoV-2 induced lung inflammation. AdCOVID and control vaccinated K18-hACE2 mice were challenged with 2.0E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020 on day 32. Mice were euthanized on DPI +4 (n = 4/group) and evaluated for lung inflammation and damage. (a) Total lung pathology score representing airway, alveolar, and vascular changes associated with inflammation, cellular hyperplasia and damage. Individual points represent the total pathology score/mouse (See Table S1 for individual scores per mouse/measurement). (b, c) Representative lung sections stained with hematoxylin and eosin and imaged at 40X (left; scale bar, 500 µm) and 400X (middle & right; scale bar, 50 µm). Bars in panel a represent mean ± SEM. Statistical analysis (n = 4 mice/group) was performed with an unpaired Student’s t test. ***, P < .001.
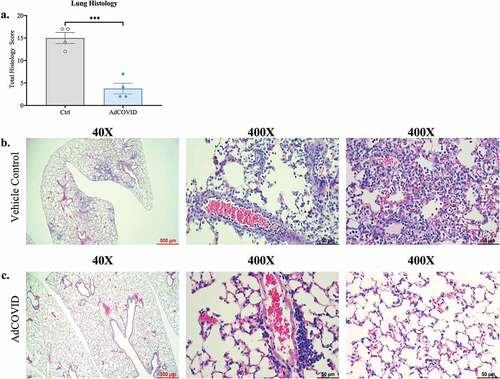
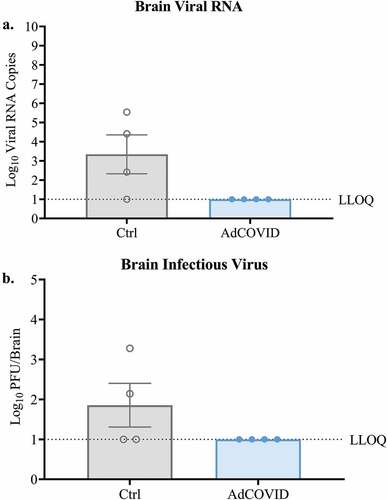
Figure 5. AdCOVID prevents neuroinvasion following intranasal SARS-CoV-2 challenge. K18-hACE2 mice were challenged with 2.0E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020 on day 32. On DPI +4, mice (n = 4/group) were euthanized, and the brains were harvested. (a) Total viral N RNA was quantified by qRT-PCR. Results are expressed as RNA copies. (b) Infectious virus was quantified in the whole brain by plaque assay. Bars represent mean ± SEM. Lowest limit of quantification (LLOQ). Statistical analyses were performed with unpaired Student’s t tests.