Figures & data
Figure 1. Illustration explaining various traits essential for the identification of NUE genotypes and different OMICS for the functional characterization of genes controlling NUE in rice.
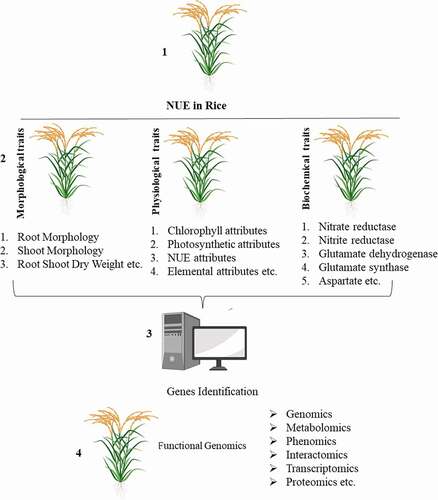
Table 1. A list of genes available in rice genome controlling NUE
Figure 2. Genes/Gene families involved in plant NUE (modified from.Citation31,Citation40

Figure 3. The basic flow chart of genome editing scheme for rice NUE improvement. (1) Selection of desirable germplasm. (2) The extraction of genomic DNA from selected germplasm. (3) Primarily analysis of genome through bioinformatics techniques to identify genes controlling NUE. (4) Selection of gene/genes of interest identified through bioinformatics analysis, available literature/online database. (5) Selection of target site based on GETs and availability/selection of vector. (6) Construction of vector holding gene of interest/target site. (7) Vector transformation through different transformation techniques (protoplast, agrobacterium transformation, and particle bombardment etc. (8) Utilization of Cas genome engineering machinery for targeted modification and extraction of genomic DNA from transgenic plants for mutation identification analysis. (9) The utilization of designed primers for PCR amplification of the target gene site to get Sanger sequencing results. (10) Screening of transgenic mutant plants based on Sanger sequencing results (type of mutation) and phenotypic changes. (11) Selection of transgene-free mutant plants for further collection of (morphological, physiological and biochemical) phenotypic data and interpretation of results.
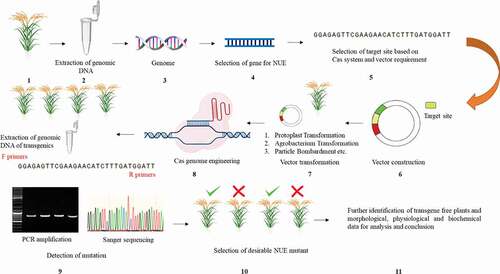
Table 2. Transgenic approaches manipulating genes controlling amino acid metabolism and transport to improve nitrogen use efficiency in rice
