Figures & data
Table 1. Enzymes used in this study.
Figure 1. Zoom view of enzyme-bound NAD (8-iodo-NAD for yADH). A. TmMtDH, B. TmGlyDH, C. yADH, D. LmG6PDH, E. rLDH, F. bGDH. PDB numbers are listed in . Several 3D models of TmMtDH were generated using Modeler softwareCitation8 and the online homology modeling server I-TASSER.Citation9-11 The different modeling approaches used single and multiple templates that each showed over 25% identity and below 10% gaps in alignments with TmMtDH. Models were analyzed using the scoring methods DOPE, DFIRE, and OPUS.Citation12-14 The best model was produced by I-TASSER using the structures of the silverleaf whitefly sorbitol dehydrogenase (PDB # 1E3J), human sorbitol dehydrogenase (PDB # 1PL8), Sulfolobus solfataricus glucose dehydrogenase (PDB # 2CDC), Thermus thermophilus threonine 3-dehydrogenase (PDB # 2DQ4), and mouse class II alcohol dehydrogenase (PDB # 1E3I) as templates. The structure was minimized using the CHARMM force fieldCitation15 and NAD was imported into TmMtDH's active site using the coordinates of NAD in human sorbitol dehydrogenase. Enzyme surfaces were visualized using The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC.
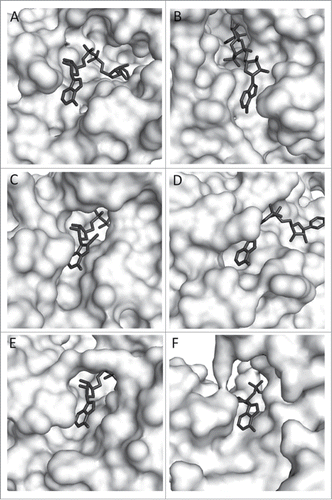
Table 2. Specific activity of the selected enzymes with 100 μM NAD and N6-CM-NAD and enzyme ranking by polarity/charge and openness of the area surrounding NAD's N6-amine. Specific activities were tested at 25°C by following the increase in A340nm in quartz cuvettes containing 100 mM substrate (L-lactate, glucose-6-phosphate, L-glutamate, or ethanol), 100 μM NAD or N6-CM-NAD in 50 mM sodium phosphate (pH 7.5). Reactions were started by adding 1.25 μg (rLDH), 1.08 μg (yADH) 0.201 μg (LmG6PDH), or 4.22 μg (bGDH) enzyme. Using the crystal structures or 3D models of the enzymes visualized in PyMOL, amino acid side-chains and backbone groups within 5 Å of the 6N amine were assessed for polarity and charge. The N6-amine solvent-accessible area was calculated using PyMOL.
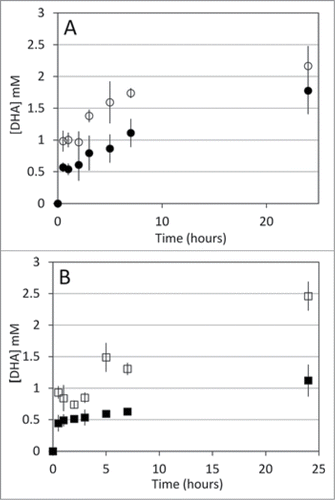
Figure 2. A. DHA accumulation during recycling reactions with yADH (○), and bGDH (•). B. DHA accumulation during recycling reactions with rLDH (□) and TmMtDH (▪). Glycerol oxidation to DHA by TmGlyDH was used to regenerate NADH in the recycling reactions. Recycling reactions were set at 25°C for all mesophilic enzymes tested and at 50°C for TmMtDH. Reactions contained 200 mM glycerol (substrate for TmGlyDH) and 100 mM acetaldehyde, pyruvate, α-ketoglutarate and NH4Cl, or fructose , as well as 45 mg Sepharose-N6-CM-NAD in 50 mM sodium phosphate (pH 7.5) (yADH, bGDH, and TmMtDH) or 50 mM Tris-HCl (pH 7.5) (rLDH). Seventeen units (one unit = amount of enzyme required to produce 1 μmol of NAD(H) per min) of TmGlyDH were used to regenerate NADH for the mesophilic enzymes (8.5 units each), and 8.5 units TmGlyDH were used to regenerate NADH for TmMtDH (4.25 units). One mM ADP was added to the bGDH reaction to activate the enzyme. Samples were collected at increasing time points over a 24 hour period, and products were quantified using a Breeze high-performance liquid chromatograph (Waters, Milford, MA) equipped with an Aminex-87C carbohydrate analysis column (Bio-Rad, Hercules, CA).