Figures & data
Figure 1. Partial amino acid sequence alignment of InuAHJ7 with GH 32 exo-inulinases. Sequence names are shown with accession numbers, except InuAHJ7, as follows: exo-inulinases from A. awamori var. 2250 (1Y4W),Citation10 Paenibacillus polymyxa ZJ-9 (AHN08014),Citation21 Geobacillus stearothermophilus KP1289 (BAC45010),Citation15 and Penicillium sp. TN-88 (BAC16218).Citation22 Identical residues are shaded in black and conserved residues are shaded in gray. The black bars indicate blocks used for designing degenerate primers. The asterisks show the putative catalytic residues.
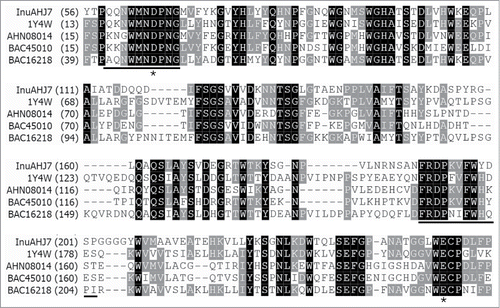
Figure 2. SDS-PAGE analysis of rInuAHJ7. Lanes: 1, cell extract from an induced transformant harboring the empty plasmid pEASY-E1; M, low-molecular weight marker; 2, crude rInuAHJ7; 3, 4, 5, and 6, rInuAHJ7 purified by Ni2+-NTA chelating affinity chromatography with imidazole gradient of 100, 200, 300, and 500 mM, respectively. The arrow indicates the band cut for MALDI-TOF MS analysis.
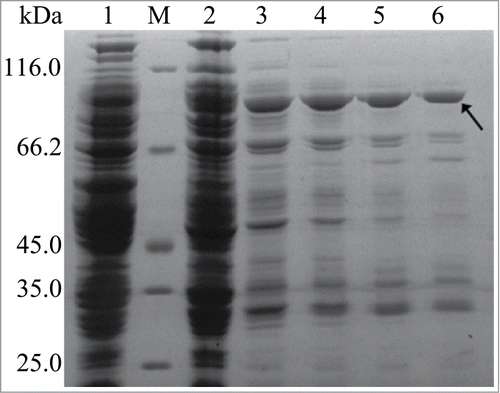
Figure 3. Characterization of rInuAHJ7. (A) Effect of pH on rInuAHJ7. (B) pH stability of rInuAHJ7. (C) Thermoactivity of rInuAHJ7. (D) Thermostability assay. The error bars represent the mean ± SD (n = 3).
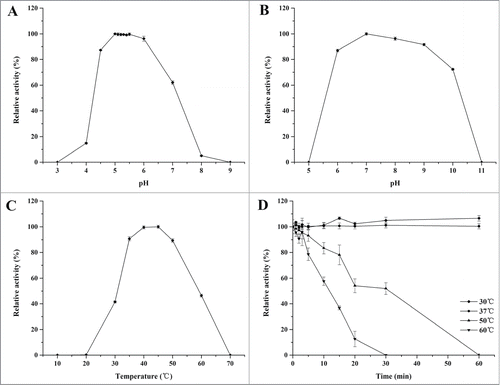
Table 1 Primers used in this study
Table 2 Effect of metal ions and organic reagents on the activity of rInuAHJ7.
Figure 4. Effect of NaCl on the activity of rInuAHJ7. The error bars represent the mean ± SD (n = 3).
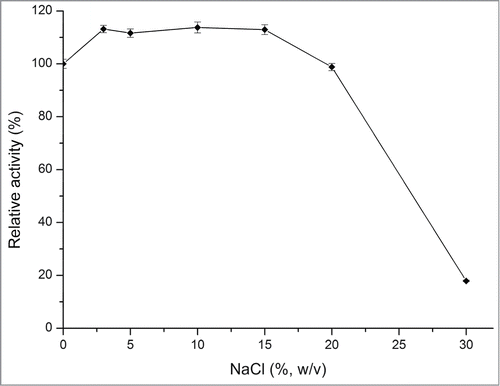
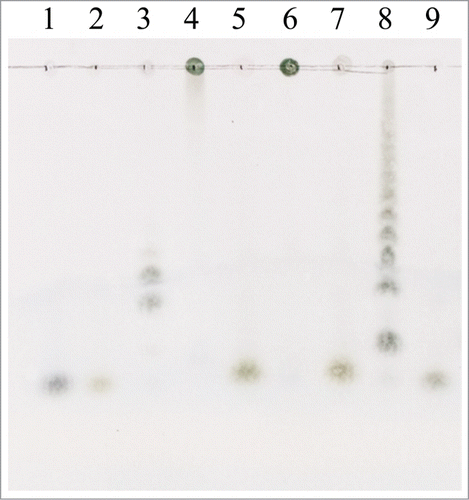
Figure 5. TLC chromatograms of the hydrolysis products of 0.5% (w/v) inulin, 0.5% (w/v) levan, and 10.0% (w/v) Jerusalem artichoke tuber powder. Lanes: 1, glucose; 2, fructose; 3, fructo-oligosaccharide mixture (kestose, nystose, and fructofuranosylnystose); 4, 6, and 8, inulin, levan, and artichoke powder with the inactivated (90°C for 5 min) rInuAHJ7, respectively; 5, 7, and 9, inulin levan, and artichoke powder hydrolyzed by rInuAHJ7 for 3 h, respectively.