Figures & data
Figure 1. Amino acid sequence alignment of Laz and Paz using the CLUSTALW program (http://clustalw.genome.ad.jp/). Laz, Neisseria gonorrhoeae azurin; Paz, Pesudomonas aeruginosa azurin. Identical and similar amino acid residues in Laz and Paz are denoted by asterisks and dots, respectively. Letters in italic and bold indicate the signal peptide and H.8 epitope, respectively. The amino acid residues responsible for binding to the metal ion are boxed. The regions corresponding to cylinders and arrows represent α-helices and β-strands, respectively. The H.8 epitope is intrinsically disordered. Laz used in this study has Ala18 instead of Cys18.
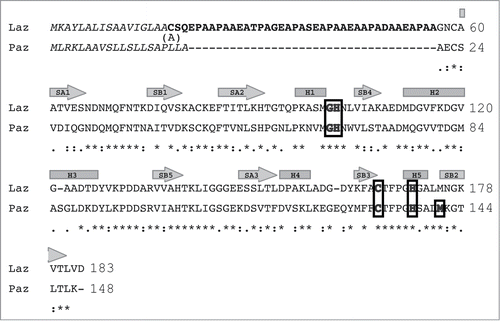
Figure 2. Molecular mass of Laz from positive-mode matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Inset, SDS-PAGE profile, lane M, molecular weight standards; lane 1, the purified Laz (10 μg).
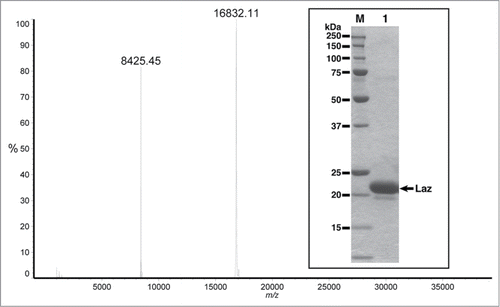
Table 1. Data collection and refinement statistics
Figure 3. X-ray crystallography of Laz. Diffraction image of the Laz crystal (inset) collected at up to 1.90 Å resolution.
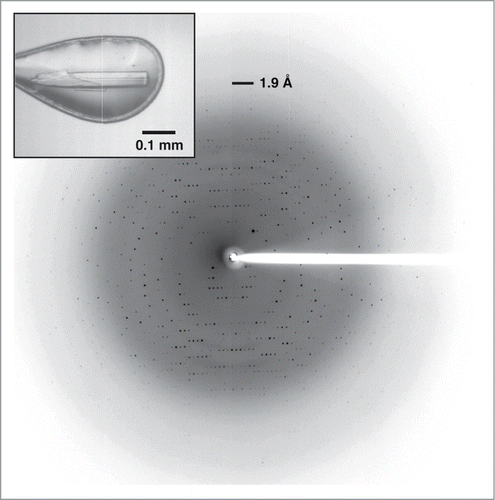
Figure 4. Structure of Laz. (A) Overall structure (stereo diagram). (B) Topology diagram. α-Helices are shown as orange cylinders and β-strands as green arrows. The metal ion is depicted by a red ball.
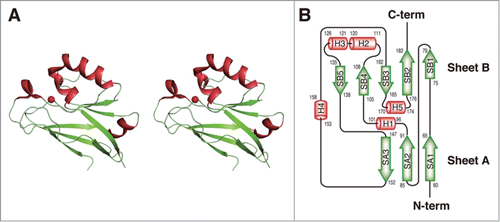
Table 2. Structure-based homology with Laz
Figure 5. Structural comparison. (A) Superimposition of the overall structures of Laz-C (green) and Paz (blue) (PDB code, 3AZU). (B) Metal-binding site. Left, Laz (carbon, green; nitrogen, blue; oxygen, red; and sulfur, yellow); right, superimposition of copper (blue)- and zinc (purple)-binding Paz proteins. Balls colored red and orange indicate the zinc and copper ions, respectively.
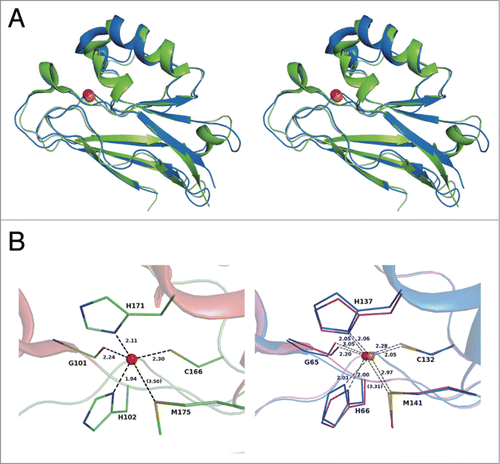
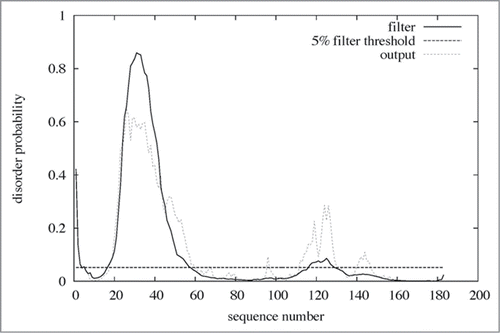
Figure 6. Disordered region in Laz. The amino acid sequence of Laz (183 residues) was analyzed by the disorder prediction program DISOPRED 2 (http://bioinf.cs.ucl.ac.uk/disopred/). Residues (20–58) corresponding to the H.8 epitope were identified as a disordered region.