Figures & data
Figure 1. Two-dimensional gel electrophoresis (2-DE) analysis of mAb H10. (A) Coomassie stained 2-DE of mAb H10 produced in agroinfiltrated N. benthamiana leaves. 800μl of protein A purified antibody (at 1mg/ml concentration) was separated by isoelectrofocusing (IEF) at pH 3-11 Non Linear (NL), followed by SDS-12.5% (w/v) PAGE. The major visible spots are indicated by numbers. Spots 7 and 10 (bold) were further characterized by N-terminal sequencing analysis. Western blot analysis of purified mAb H10 separated by 2DE using anti-LC (B) or anti-HC (C) specific antibodies. Spot numbers in brackets indicate the correspondence (based on observed pI and molecular weight values) to the spots on Coomassie stained gel of Figure 1A.
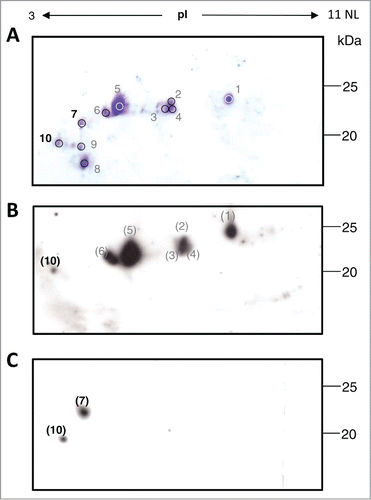
Figure 2. N-terminal sequence analysis of spots 7 and 10 and Heavy chain (HC) cleavage sites of mAbs 2G12 and H10. (A) Summary of results obtained by N-terminal sequence analysis of spots 7 and 10. Top row: domain architecture of complete LC and HC antibody sequences. Middle and bottom rows: the N-terminal sequences of mAb H10 fragments in spots 7 and 10 is reported, together with the deduced molecular species. n.d.: N-terminal sequences in that region were not detected. (B) Alignment of mAbs H10 and 2G12 HC amino acid sequences. The variable (VH) and constant (CH1, CH2 and CH3) domains and hinge region of the heavy chain are indicated and separated by black vertical bars. Open arrows indicate cleavage sites of mAbs 2G12 and H10 previously identified by Hehle and colleagues,Citation11 filled arrows indicate mAb 2G12 cleavage sites identified by Niemer and colleagues.Citation14 The newly identified cleavage site in mAb H10, at the VH-CH1 interface, is indicated by the black diamond-arrow sign and the STKGP N-terminal sequence deriving from it is underlined.

