Figures & data
Figure 1A. A schematic of the designed microfluidic instrument. Flow through the tubing is generated using the syringe pump. The robotic stage moves into the bacteria sample to develop droplets. To measure the optical density a photodiode was embedded into a the incubation plate.
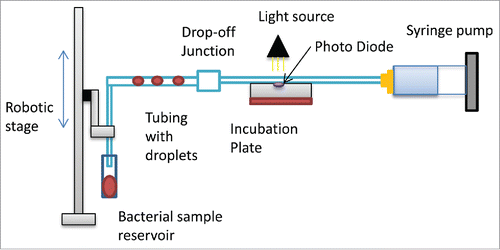
Figure 1B. Images of drop-off junction mixing system where the small droplet is a drug droplet and the large pink droplet is a bacteria culture droplet. Droplets move from a smaller diameter tube into a larger diameter tube. Within the larger tube the droplets are smaller than the diameter of the tubing they move together and coalesce to form a fully mixed droplet and then moves back to 812um tubing for incubation.

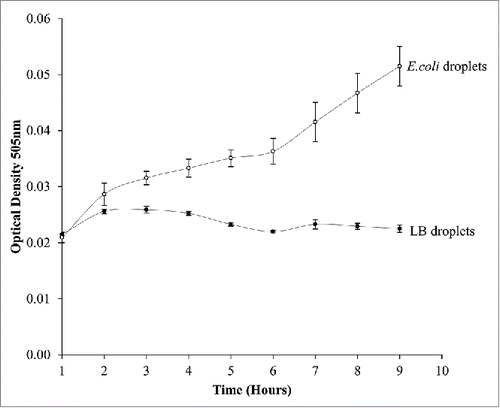
Figure 2. A graph comparing growth rates of traditional E.coli shaker flask culture with E.coli droplet cultures measured on a microplate reader. Droplet cultures show a higher growth rate than orbital shaker cultures.
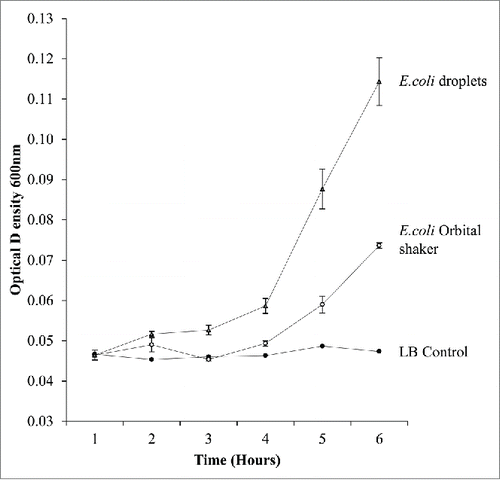
Figure 4. An image of the voltage readings from the photodiode system. Each peak represents a droplet where the first 10 droplets are LB followed by 10 droplets of E.coli readings taken after 6 hours of incubation.
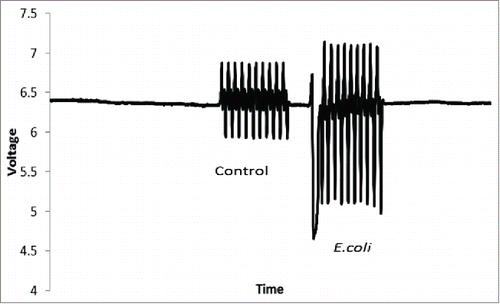
Figure 5. Monitoring the effect of variation of the droplets volume (600 nl to 2500 nl) on bacterial growth over time. Results indicated that the optimum growth rate is achieved in 1500nl droplet volume
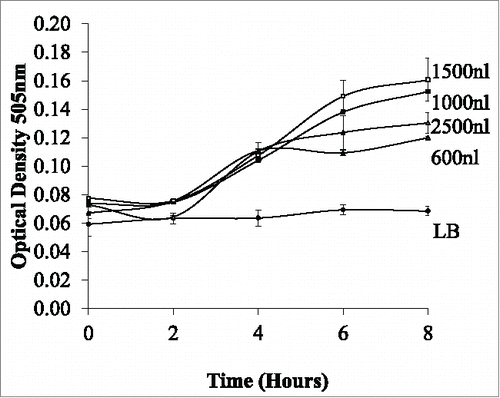
Figure 6. Monitoring the effect of dynamic conditions on E.coli growth within a droplet at varying flow rates from 0-60ul/min. Increasing the flow rate increases the growth rate up to 30ul/min.
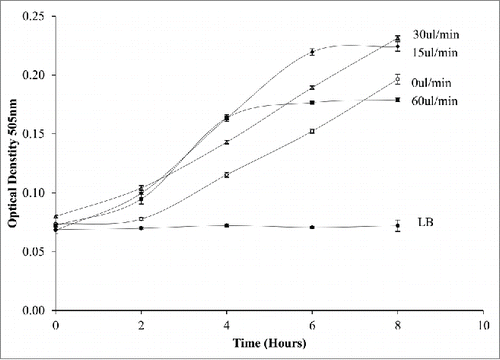
Figure 7A. A diagram of the internal circulations within a droplet where there are 2 regions of circulation; center rotations and 2 rotations at each cap end. The line demonstrates the region where most of the cells reside.
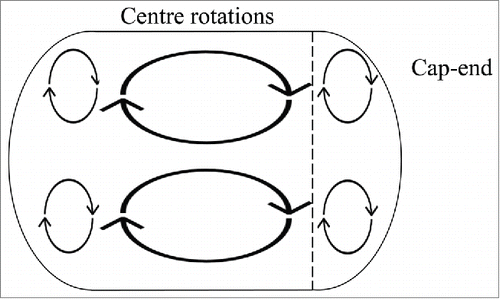
Figure 7B. Images of the bacteria droplets at varying flow rate 15–60 ul/min. A dotted line shows the area of most concentration at 15 ul/min, this line is placed at the same point in shown in 30 ul/min and 60ul/min where it is clear that the bacteria moves
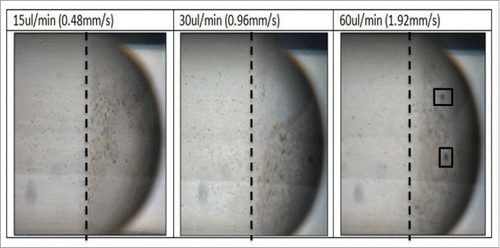
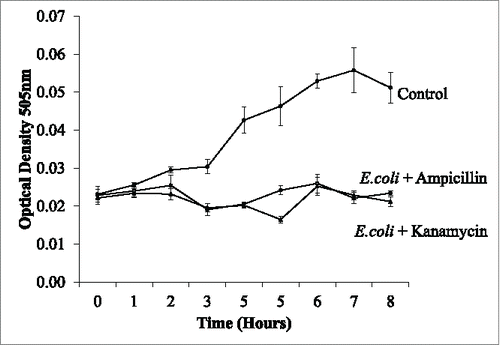
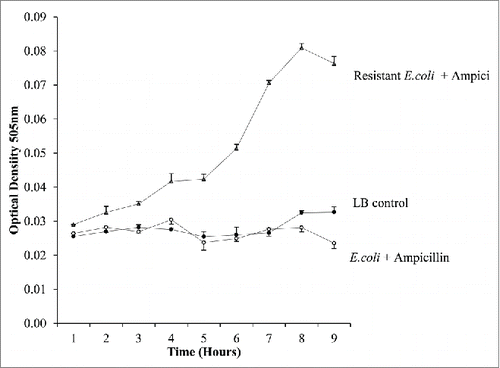
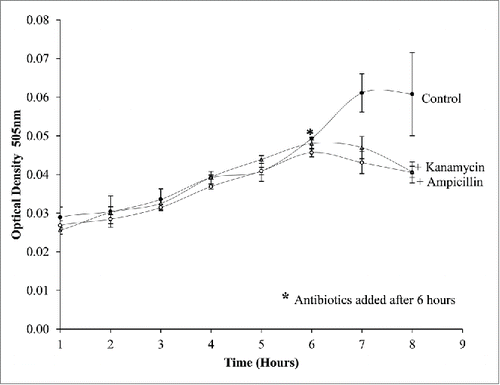
Figure 8. The treatment of bacterial cultures within the droplets. Droplets of; E.coli, E.coli mixed with ampicillin and E.coli mixed with Kanamycin were monitored over time. Antibiotic treated droplets are prevented from growing while the non-treated E.coli continue to grow further into the cap ends. At 60 ul/min 2 internal circulations are visibly present in the cap ends.