Figures & data
Figure 1. Lignin subunits attacked by enzymes and their most common reaction mechanisms. The bulky lignin polymer structure represents part of an organosolv lignin substrate.Citation65 Both superfamilies of lignin-degrading enzymes (heme peroxidases and laccases) can oxidize phenolic lignin subunits. In order to oxidize non-phenolic lignin subunits, laccase requires the presence of a mediator. Lignin peroxidase, versatile peroxidase and DyP-type peroxidase do not require a mediator to attack non-phenolic structures. Manganese peroxidase and versatile peroxidase oxidize phenolic lignin subunits via the oxidation of manganese (Mn2+ → Mn3+). The inset box shows heme peroxidases reducing H2O2 to water to catalyze the oxidation reactions, whereas laccases reduce molecular oxygen to water, which is accompanied by the oxidation of the substrates or mediators. The atoms and bonds of the phenolic and non-phenolic lignin subunits are highlighted in bold within the bulky structure. Abbreviations: med = mediator, sub = substrate.
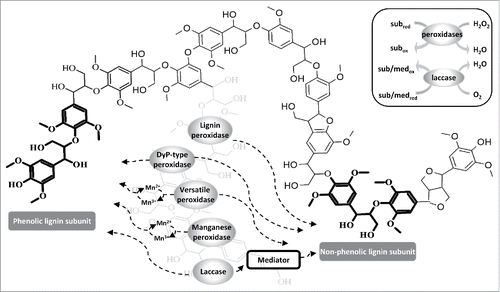
Table 1. Recombinant fungal lignin-degrading peroxidases. Examples of successfully produced fungal lignin-degrading peroxidases and the substrates that have been used. The activity of the enzymes (if stated) is shown in parentheses. ABTS: 2,2-azinobis (3-ethylbenzothiazoline-6-sulfonate), DCP: 2,4-dichlorophenol, DFAD: 4-[(3,5-difluoro-4-hydroxyphenyl)azo]benzenesulfonic acid, DMP: 2,6-dimethoxyphenol, icb: inclusion body; MHG: methoxyhydroquinone, VA: veratryl alcohol.
Table 2. Recombinant bacterial lignin-degrading peroxidases. Examples of successfully produced recombinant bacterial lignin-degrading peroxidases and the substrates that have been used. The activity of the enzymes (if stated) is shown in parentheses. ABTS: 2,2-azinobis (3-ethylbenzothiazoline-6-sulfonate), DCP: 2,4-dichlorophenol, DMP: 2,6-dimethoxyphenol, DOPA: L-3,4-dihydroxyphenylalanine, VA: veratryl alcohol.
