Figures & data
Table 1. Strains used in this study.
Table 2. Oligonucleotides used in this study.
Figure 1. Construction of deletion cassette with long flanking homology regions by overlapping PCR. In the first round of PCR, the upstream sequence of the nth1 gene, the kanr marker gene and the downstream sequence of the nth1 gene were amplified in 3 different PCR reaction tubes. S. cerevisiae genomic DNA was used as template for amplification of 0.26 kb 5′ UP nth1 region and 0.4 kb 3′ DOWN nth1 region by using primers L1, L2, and L3, L4, respectively. The pFA6a-kanMX6 vector was used as a template for amplification of kanMX region by using primers kanMX-F and kanMX-R. In the second round of PCR, the 5′ UP nth1 region and kanMX region were fused together by using primers L1 and kanMX-R. In the third round of PCR, the 3′ DOWN nth1 region and second round PCR product (5′ UP nth1 region-kanMX region) were fused together by using primers L1 and L4. The third round PCR product (5′ UP nth1 region- kanMX region - 3′ DOWN nth1 region) was cloned into pGEMT vector for construction of the disruption cassette. After transformation of the disruption cassette into S. cerevisiae, homologous recombination takes place with the deletion of target gene.
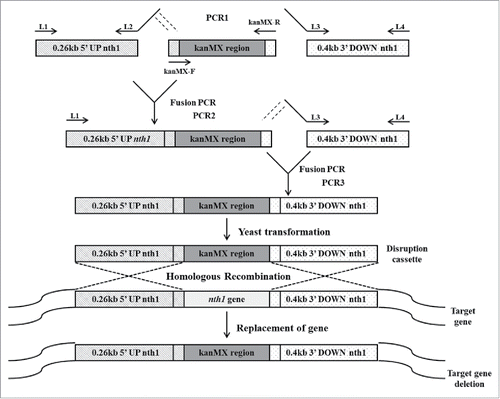
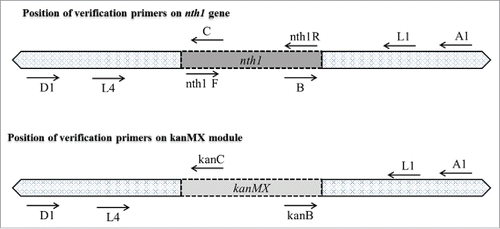
Figure 3. Verification of gene deletion strain. Genomic DNA of wild strain, heterozygous strain, and homozygous strain (Agarose gel image A, B, and C, respectively) were used as templates for PCR with different sets of primers. In all of the gels, Lane M: Gene-Direx 100 bp DNA Ladder, lane 1: primers Nth1-F and R, lane 2: primers L1 and L4, lane 3: primers A1 and kan B, lane 4: primers A1 and B, lane 5: primers D1 and kan C, lane 6: primers D1 and C.
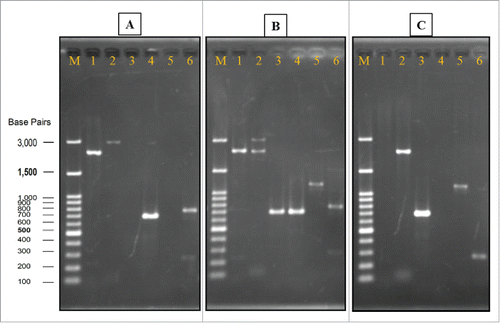
Figure 4. PCR confirmation of the recombinant expression vector pGAPZC-tps1 (A) and Over-expression of TPS (B) in SC (wild strain), SCT (tps1 overexpression strain) and SCTΔN (tps1 overexpression and nth1 deletion strain).
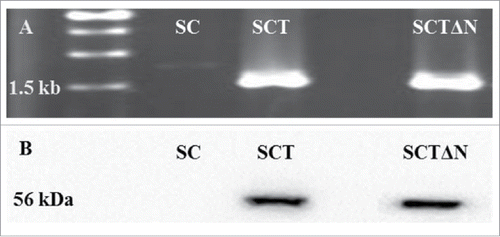
Table 3. Intracellular trehalose content of yeast cells during growth and under stress.
Figure 5. Effect of ethanol concentrations on 24 h growth of wild strain (SC) and engineered strain (SCTΔN). Control (C) is the initial OD of strains. Data are presented as the means ± SD (n = 3). *Significantly higher than the other group. One-way ANOVA, Student's t test, P < 0.05. Bar values with the different letters (A, B, C, D, E, F for SC, while a, b, c, d, e, f, g for SCTΔN) are significantly different. One-way ANOVA, Duncan's multiple range test, P < 0.05.
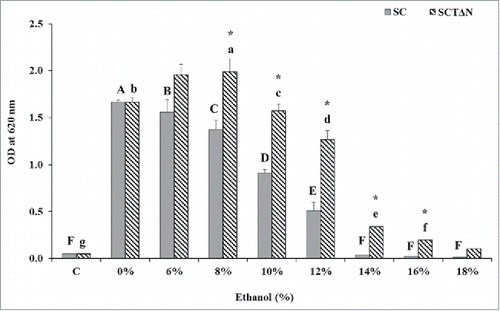
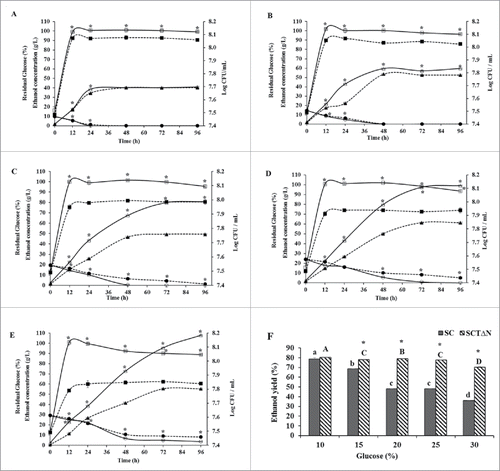
Figure 6. Ethanol production capacities of wild strain (SC) and engineered strain (SCTΔN). Ethanol concentration in g/L (triangle), biomass Log CFU/mL (square) and residual glucose concentration in % (circle) of SC (wild strain) (closed) and SCTΔN (tps1 overexpression and nth1 deleted strain) (open) grown in YP broth (10 g/L yeast extract, 20 g/L peptone) containing different concentrations of glucose. A: 10% glucose; B: 15% glucose; C: 20% glucose; D: 25% glucose; E: 30% glucose; F: Ethanol yield (%) after 96 h of incubation. Data are presented as the means ± SD (n = 3). *Significantly higher than the other group. One-way ANOVA, Student's t test, P < 0.05. Bar values of the ethanol yield (%) with the different letters (a, b, c, d for SC and A, B, C, D for SCTΔN) are significantly different. One-way ANOVA, Duncan's multiple range tests, P < 0.05.