Figures & data
Figure 1. LINC00460 knockdown inhibited the cell growth of SW1990 cells. (a–b) The LINC00460 mRNA expression was quantified by qRT-PCR. (c) The cell viability of SW1990 cells was analyzed by CCK8 assay. (d) The cell proliferation of SW1990 cells was analyzed by colony formation assay. Error bars represent the mean ± SEM from three independent experiments. **P< 0.01, ***P< 0.001 vs. Control
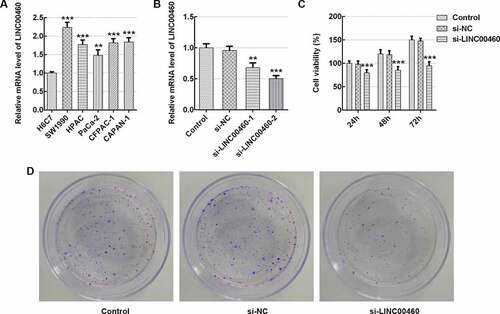
Figure 2. LINC00460 knockdown inhibited the migration of SW1990 cells. (a–b) The migration of SW1990 cells transfected with si-LINC00460 or si-NC was detected by wound-healing assay. Error bars represent the mean ± SEM from three independent experiments. ***P< 0.001 vs. Control

Figure 3. LINC00460 knockdown inhibited the invasion of SW1990 cells. (a–b) The invasion of SW1990 cells transfected with si-LINC00460 or si-NC was detected by transwell chamber assay. Error bars represent the mean ± SEM from three independent experiments. ***P< 0.001 vs. Control
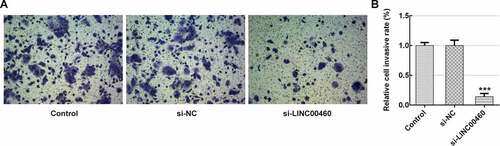
Figure 4. LINC00460 knockdown promoted the apoptosis of SW1990 cells. (a–b) The apoptosis rate of SW1990 cells was detected by flow cytometry. (c) The expressions of proteins including Bcl-2, Bax, caspase3, pro-caspase3, caspase7 and pro-caspase7 were determined by western blotting. Error bars represent the mean ± SEM from three independent experiments. **P< 0.01, ***P< 0.001 vs. Control
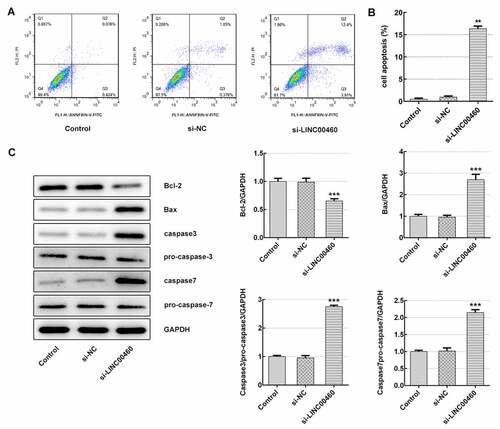
Figure 5. LINC00460 directly target miR-320b and downregualted miR-320b expression. (a) The mRNA expression of miR-320b in multiple pancreatic cancer cell lines was quantified with qRT-PCR. (b) Binding sites of LINC00460 and miR-320b predicted by Starbase website. (c) The relative luciferase activity was detected by luciferase reporter assay. (d) The mRNA expression of miR-320b in SW1990 cells transfected with si-LINC00460 or si-NC was quantified with qRT-PCR. Error bars represent the mean ± SEM from three independent experiments. *P< 0.05, ***P< 0.001 vs. Control
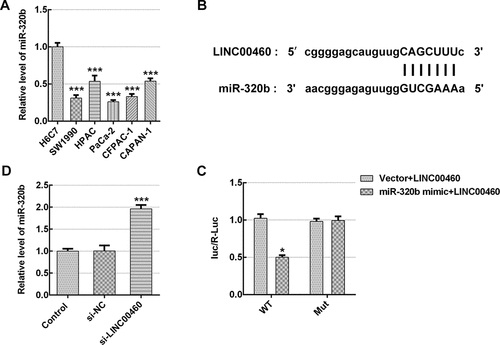
Figure 6. MiR-320b dowregulation partly reversed the effect of LINC00460 knockdown on the proliferation of SW1990 cells. (a) The mRNA expression of miR-320b in SW1990 cells transfected with miR-320b inhibitor or inhibitor-NC was quantified with qRT-PCR. (b) The cell viability of SW1990 cells was analyzed by CCK8 assay. (d) The cell proliferation of SW1990 cells was analyzed by colony formation assay. Error bars represent the mean ± SEM from three independent experiments. ***P< 0.001 vs. Control. ###P< 0.001 vs. si-LINC00460
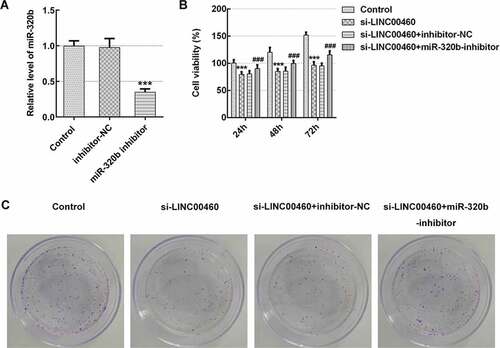
Figure 7. MiR-320b dowregulation partly reversed the effect of LINC00460 knockdown on the migration of SW1990 cells. (a–b) The migration of SW1990 cells transfected with si-LINC00460, si-LINC00460+ inhibitor-NC or si-LINC00460+ miR-320b-inhibitor was detected by wound-healing assay. Error bars represent the mean ± SEM from three independent experiments. ***P< 0.001 vs. Control. ###P< 0.001 vs. si-LINC00460

Figure 8. MiR-320b dowregulation partly reversed the effect of LINC00460 knockdown on the invasion of SW1990 cells. (a–b) The invasion of SW1990 cells transfected with si-LINC00460, si-LINC00460+ inhibitor-NC or si-LINC00460+ miR-320b-inhibitor was detected by transwell chamber assay. Error bars represent the mean ± SEM from three independent experiments. ***P< 0.001 vs. Control. ##P< 0.01 vs. si-LINC00460
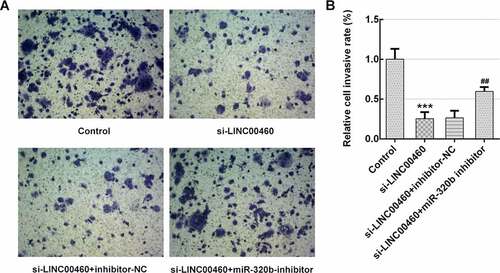
Figure 9. MiR-320b dowregulation partly reversed the effect of LINC00460 knockdown on the apoptosis of SW1990 cells. (a–b) The apoptosis rate of SW1990 cells was detected by flow cytometry. (c) The expressions of proteins including Bcl-2, Bax, caspase3, pro-caspase3, caspase7 and pro-caspase7 were determined by western blotting, and quantification (d). Error bars represent the mean ± SEM from three independent experiments. ***P< 0.001 vs. Control. ##P< 0.01, ###P< 0.001 vs. si-LINC00460
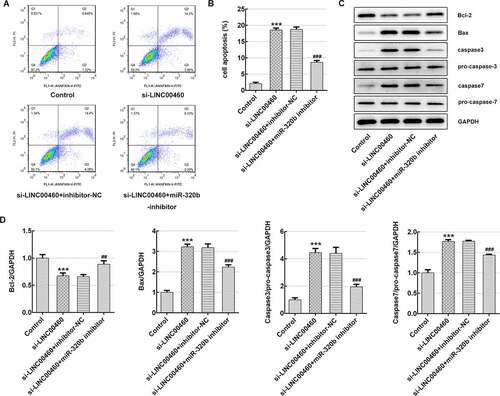
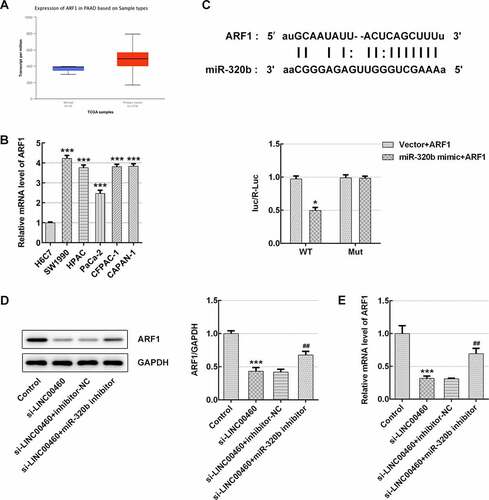
Figure 10. miR-320b/ARF1 axis mediated the effect of LINC00460 knockdown on cell growth of SW1990 cells. (a) The ARF1 level in PAAD patients by UALCAN website. (b) The ARF1 mRNA level was detected by qRT-PCR. (c) Binding sites of miR-320b and ARF1 predicted by Starbase website, and the relative luciferase activity was detected by luciferase reporter assay. (d) The ARF1 proteins expression was quantified by western blotting. (e) The ARF1 mRNA level was detected by qRT-PCR. Error bars represent the mean ± SEM from three independent experiments. *P< 0.05, ***P< 0.001 vs. Control; ##P< 0.01 vs. si-LINC00460