Figures & data
Figure 1. Schematic detailing the experimental culture routes investigated. Cells in each route were seeded according to the density stated in the figure. Following 48 h culture route A1, A3, and B1 were subjected to a 100% medium exchange. Following 72 h, all routes were passaged. Route A1, A2, A3 and A4 underwent a further two passages (experiment A). Route B1 and B2 underwent a further nine passages (experiment B); *M. Ex = medium exchange
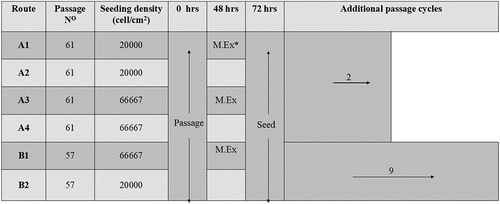
Table 1. Summary of the significance values at each passage, comparing the SMRs of the different culture routes using Tukey’s multiple comparisons test. Both seeding density and feeding regime are shown to result in significant difference between the four routes. The highest significant differences between routes, within passages, are observed when both parameters are changed, e.g. A2 vs A3 (n = 2 for each condition)
Figure 2. (a) SGR over three passages, similar values are observed in all culture routes, with the exception of A4 at passages 3 (n= 2). (b) Glucose SMR over three passages demonstrating that route conditions B2 and B4 that have no medium exchange following 48 h have higher SMR compared to route A1 and A3, significant differences between routes at each passage shown on the graph highlight that seeding density, feeding regime and a combination of both parameters result in a different SMRs (n = 2). (c) Oct3/4 marker expression over three passages, expression levels are similar between routes, significant difference observed between passages 2 and 3 for all routes (*p < 0.05) except route A4. (d) SSEA-1 marker expression over three passages, inset shows that percentage expression levels are below 1.5%. (e) SSEA-4 marker expression over three passages, significant differences are observed only within route A2 at the different passages (*p < 0.05). N.B. Oct 3/4 and SSEA-4 are positive markers for pluripotent human embryonic stems, SSEA-1 is a negative marker associated with pluripotency of human embryonic stem cells
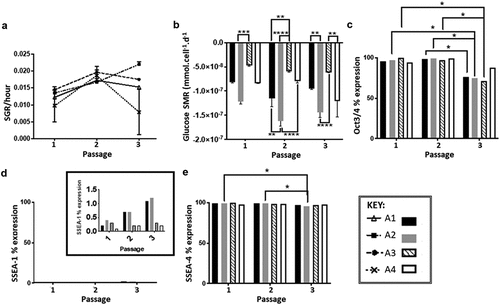
Figure 3. (a) Specific growth rate trend for route B1 and B2 over 10 passages, showing a fluctuation in SGR throughout all 10 passages that follows the same trend regardless of route, n= 3 error bars showing standard deviation, SD. (b) Cell viability trend of route B1 and B2 over 10 passages both routes follow a similar trend in viability throughout all passages cycles, route B1 had lower viability during 5 of the passage cycles (4−8) in comparison to route B2, n= 2 error bars showing SD. (c) Glucose SMR from passage cycle 2 to 10 demonstrating the significant differences between passages (***p < 0.001), at each individual passage cycle route B2 is significantly higher than route B1 (****p < 0.0001) (d) Oct3/4 marker expression percentages for experiment B at passage cycle 1, 3, 7 and 9 significant differences between passages (****p < 0.0001) and significant differences between culture route B1 and B2 (p = 0.0004, n= 3). (e) SSEA-1 marker expression percentages for experiment significant differences between culture route B1 and B2 at passages 3,7 and 9, B2 exhibited higher expression levels of SSEA-1 compared to B1. (f) SSEA-4 marker expression percentages for experiment B, no significant differences in expression levels are observed both between passages and within the different routes, B1 and B2
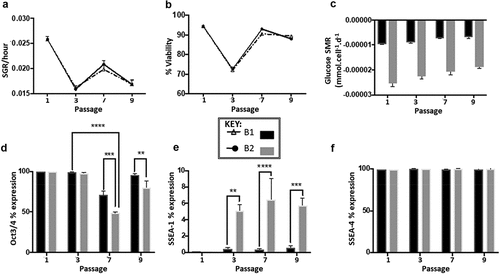
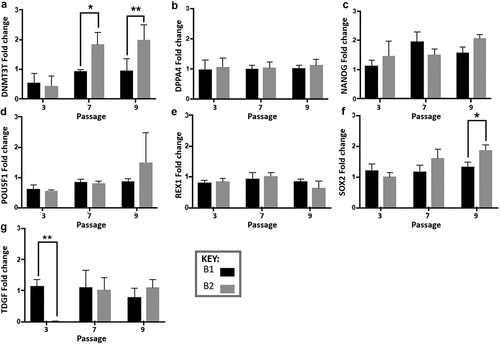
Figure 4. Gene expression analysis of experiment B at three passage points over the 10 passages (normalized to the cells at passage 1), expression was measured by real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Fold change was calculated using the relative quantity of each gene was calculated by the ΔΔCt method, using a correction for the amplification efficiency of that gene, and normalized to the geometric mean of two housekeeping genes: glyceraldehyde-3-phosphate dehydrogenase and β-actin. Significant differences in fold change at different passage cycles were seen only for A-DNMT3B (***p = 0.0007), NANOG (*p = 0.01) and SOX2 (**p = 0.004). Significant differences between route A1 and A2 were seen only for A -DNMT3B (**p = 0.0032) and F-SOX2 (**p = 0.006). Differences between the two routes within a passage cycle were only observed for G-TDGF (passage cycle 3, **p = 0.018), A-DNMT3B (passage cycle 7, *p = 0.0221; passage cycle 9, **, p = 0.0073), and F-SOX2 (passage cycle 9, *p = 0.0191). Error bars indicate standard deviation (n= 3 for all data presented). N.B. Cells from passage 1 were used as the control sample for the qRT-PCR fold change analysis, therefore no data is presented for passage 1 in the graphs