Figures & data
Table 1. 127 autophagy-related genes
Figure 1. LASSO cox-regression analysis to filter the autophagy-related gene pairs for prognosis. (a)The most representative autophagy-related gene pairs were obtained by LASSO analysis; (b) LASSO coefficient of the 13 autophagy-related gene pairs
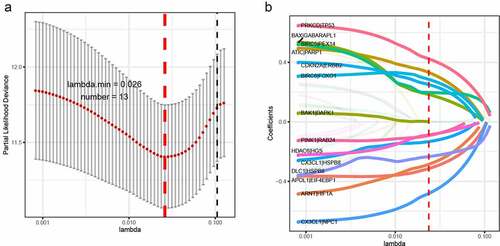
Figure 2. Survival analysis of HCC in the two cohort. (a)Risk scores distribution of patients in the training cohort; (b) Survival time and survival state of patients in the training cohort; (c) Kaplan-Meier survival curve of patients with HCC in the training cohort; (d) Risk scores distribution of patients with HCC in the validation cohort; (e) Survival time and survival state of patients with HCC in the validation cohort; (f) Kaplan-Meier analysis of patients in the validation cohort
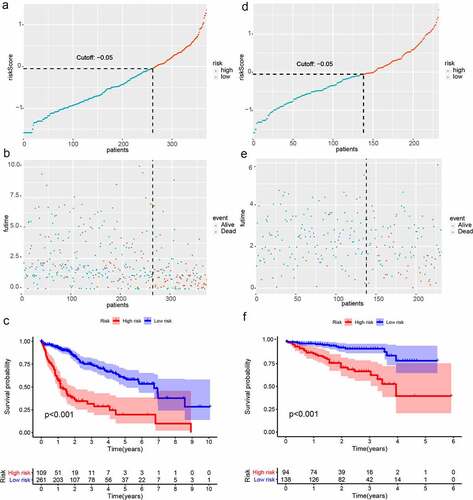
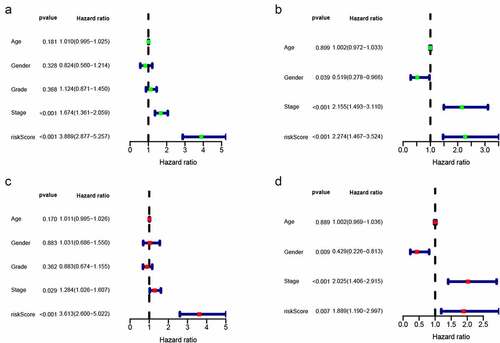
Figure 3. Univariate and multivariate analyses showed independently prognostic factors for OS of HCC in the two cohorts. (a) Univariate analyses in the training cohort; (b) Multivariate analyses in the training cohort; (c) Univariate analyses in the validation cohort; (d) Multivariate analyses in the validation cohort
Supplemental Material
Download ()Data availability statement
The datasets generated and/or analyzed during the current study are available in TCGA (https://portal.gdc.cancer.gov) and ICGC (https://icgc.org/).