Figures & data
Table 1. The interference sequences of siDUXAP8
Table 2. The sequences of primer
Figure 1. DUXAP8 expression was upregulated in PTC cell lines and DUXAP8 silencing suppressed PTC cell proliferation. (a) Expression of DUXAP8 in different PTC cell lines. (b-c) Expression of DUXAP8 in GLAG-66 and TPC-1 cells transfected with DUXAP8 siRNAs. (d-e) The proliferation of GLAG-66 and TPC-1 cells was determined by CCK-8 assay. ## p < 0.01, compared to siNC
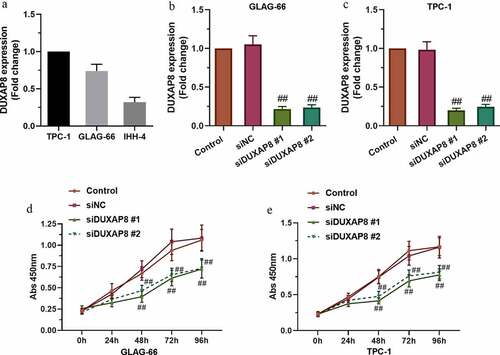
Figure 2. DUXAP8 silencing inhibited PTC cell migration and invasion. (a-b) The migration ability of GLAG-66 and TPC-1 cells was assessed using a wound-healing assay. (c-d) The invasive capacity of GLAG-66 and TPC-1 cells was detected through the transwell invasion assay. # p < 0.05, ## p < 0.01, compared to siNC
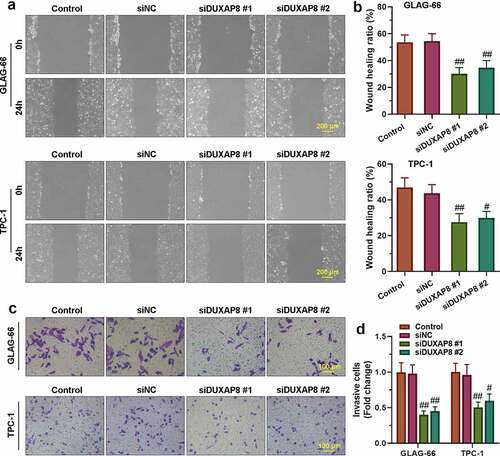
Figure 3. DUXAP8 bound to miR-223-3p and suppressed the expression of miR-223-3p. (a) Sequence alignment of DUXAP8 binding to miR-223-3p. (b) The binding activity between DUXAP8 and miR-223-3p was measured by luciferase reporter assay. ** p < 0.01. (c-d) Expression of miR-223-3p in GLAG-66 and TPC-1 cells was examined using qRT-PCR. ## p < 0.01, compared to siNC
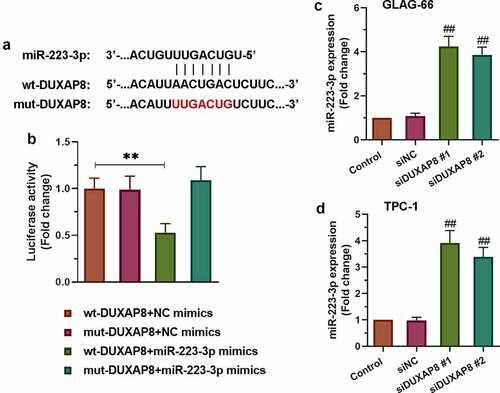
Figure 4. miR-223-3p over-expression inhibited PTC cell proliferation, migration, and invasion. (a-b) CCK-8 assay was performed to examine the proliferation of GLAG-66 and TPC-1 cells. (c-d) A wound-healing assay was carried out to measure GLAG-66 and TPC-1 cell migration. (e-f) The transwell invasion assay was used to assess GLAG-66 and TPC-1 cell invasion. # p < 0.05, ## p < 0.01, compared to NC mimics
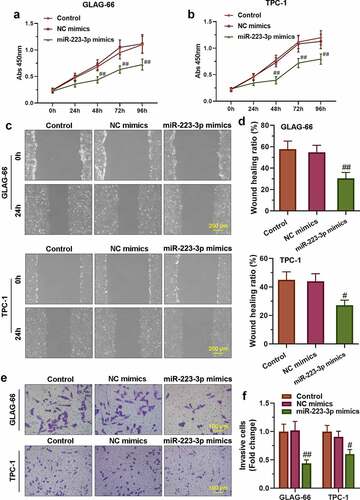
Figure 5. CXCR4 was a target of miR-223-3p. (a) Sequence alignment of CXCR4 binding to miR-223-3p. (b) The binding activity between CXCR4 and miR-223-3p was measured by luciferase reporter assay. ** p < 0.01. (c-d) Expression of CXCR4 in GLAG-66 and TPC-1 cells was examined using qRT-PCR. ## p < 0.01, compared to NC mimics
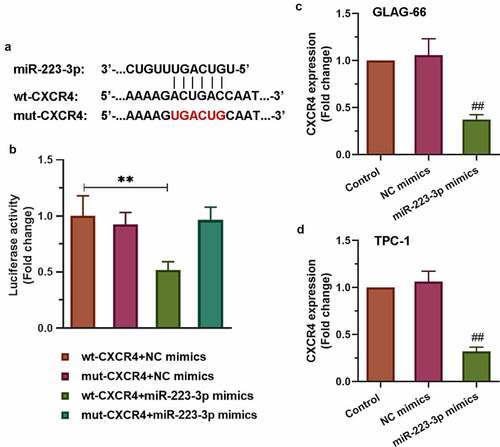
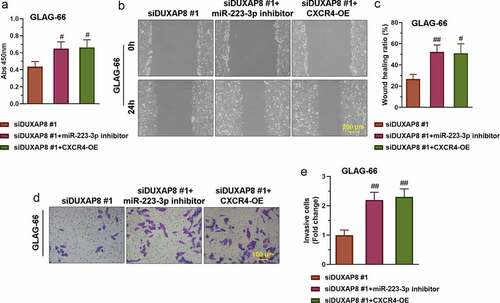
Figure 6. DUXAP8 silencing suppresses PTC cell proliferation, migration, and invasion via the miR-223-3p/CXCR4 axis. (a) The proliferation of GLAG-66 cells was tested using CCK-8. (b-c) The migration activity of GLAG-66 cells was detected by the wound healing assay. (d-e) The invasion of GLAG-66 cells was measured by the transwell invasion assay. # p < 0.05, ## p < 0.01, compared to siDUXAP8 #1
Supplemental Material
Download ()Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request. The supplemenary data were aquired from the database (http://gepia.cancer-pku.cn/detail.php?gene=DUXAP8).