Figures & data
Table 1. Primers in this study
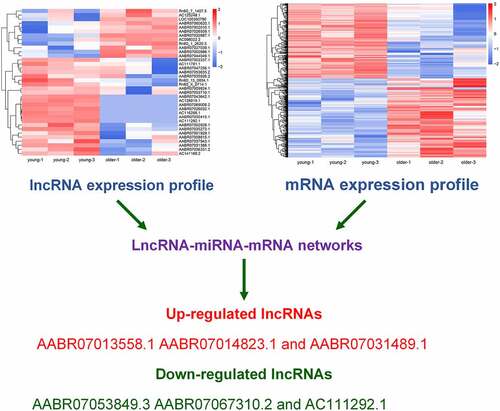
Figure 1. Differential lncRNAs profiles in thoracic aortas of young and older rats. (a) A Venn diagram showing the numbers of lncRNAs characterized in the young and older rat’ thoracic aortas. Using RNA deep sequencing, a total of 269 common lncRNAs were identified in each group. (b) The size distribution of lncRNAs identified in the rat’ thoracic aortas by RNA sequencing. (c) The genomic location of lncRNAs significantly differentially expressed in the thoracic aortas of young and older rats. Upregulated and downregulated lncRNAs in older rats compared to those of young were shown in red and green colors, respectively. (d) Hierarchical clustering of differentially expressed lncRNAs between the young and older rat’ thoracic aortas. The alterations in the expression of lncRNAs are presented as heatmaps, in which high and low expressions of lncRNAs were shown in red and blue colors, respectively. lncRNAs: long non-coding RNAs; chr: chromosome
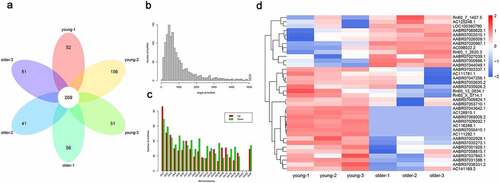
Figure 2. Significantly altered mRNA profiles in thoracic aortas of young and older rats. (a) Hierarchical clustering of differentially expressed mRNAs in the thoracic aortas of the young and older rats. mRNAs with high and low expression are shown in red and blue colors, respectively. (b) A scatter plot presenting the expression changes in mRNAs of the young and older rats’ thoracic aortas. mRNAs with upregulated, downregulated, and similar expressions in the young rats compared with older ones are shown in red, green, and blue colors, respectively. DEG: differentially expressed genes; TPM: transcripts per million
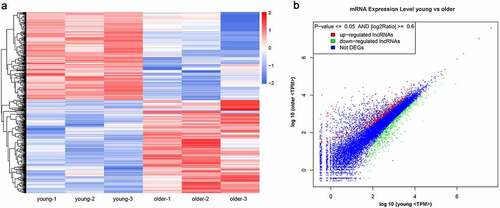
Figure 3. Functional categorization of significantly altered mRNAs between young and older rats’ thoracic aortas. (a) Categorization of differentially expressed mRNAs between young and older rat’ thoracic aortas using GO terms. (b) The KEGG enrichment signaling pathways of differentially expressed genes between young and older rat’ thoracic aortas. GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes
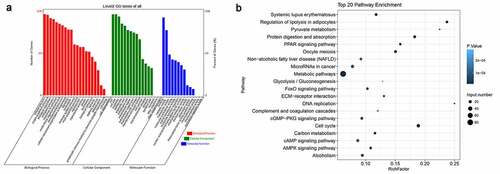
Figure 4. lncRNA–microRNA–mRNA networks. lncRNA–microRNA–mRNA networks were constructed using Cytoscape software base on the differentially expressed lncRNAs and mRNAs identified in the young and older rat’ thoracic aortas by RNA deep sequencing. LncRNAs, microRNAs, and mRNAs are indicated by red diamonds, yellow irregular quadrilaterals, and pink ovals, respectively. Predicted interactions between them are shown by gray lines. The mRNAs involved in collagen synthesis are highlighted with green arrow. Detailed information is provided in Supplemental Table 2
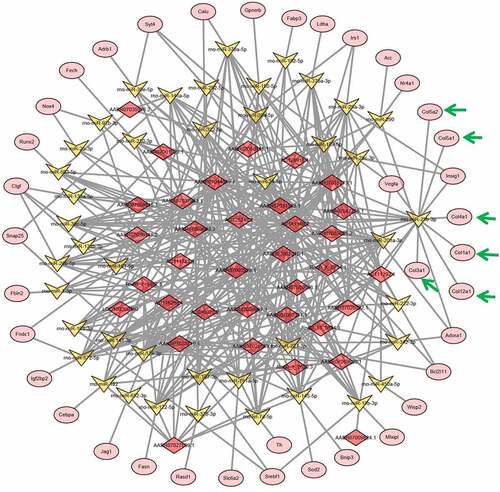
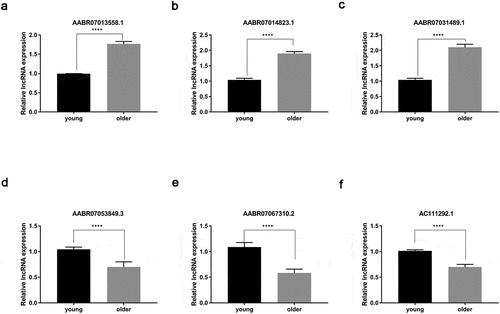
Figure 5. Verification of differential expression of representative lncRNAs between young and older rat’ thoracic aortas. The upregulated expression levels of AABR07013558.1 (a), AABR07014823.1 (b), and AABR07031489.1 (c) as well as the downregulated expression of AABR07053849.3 (d), AABR07067310.2 (e), and AC111292.1 (f) in the thoracic aortas of older rats compared to young ones were assessed using the qRT-PCR method. At least three biological replicates were performed, GAPDH as the internal standard. lncRNAs: long non-coding RNAs; ****P < 0.0001