Figures & data
Table 1. Primer sequence
Table 2. Correlations between circ_0000467 expression and clinical characteristics in CRC patients
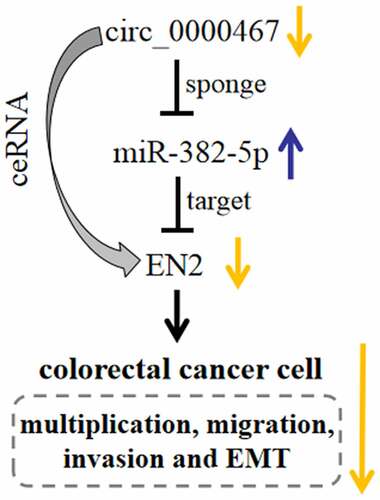
Figure 1. The expression of circ_0000467 was up-modulated in CRC
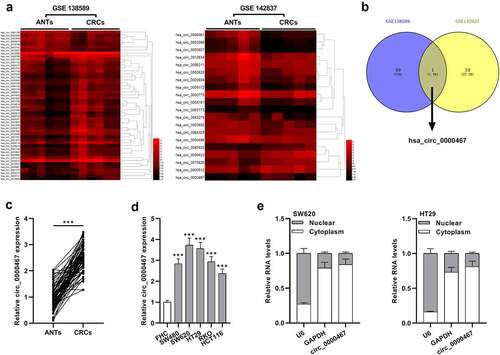
Figure 2. Knockdown of circ_0000467 impeded the multiplication, migration, and invasion of CRC cells
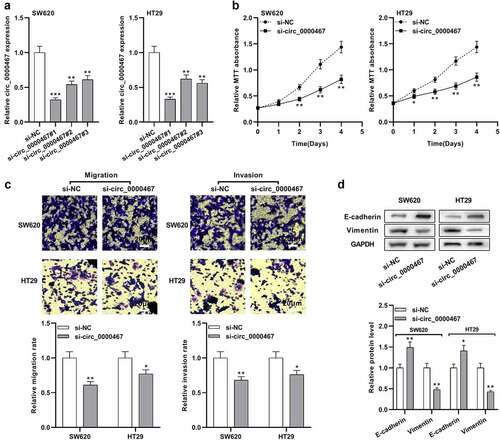
Figure 3. MiR-382-5p was a downstream target of circ_0000467
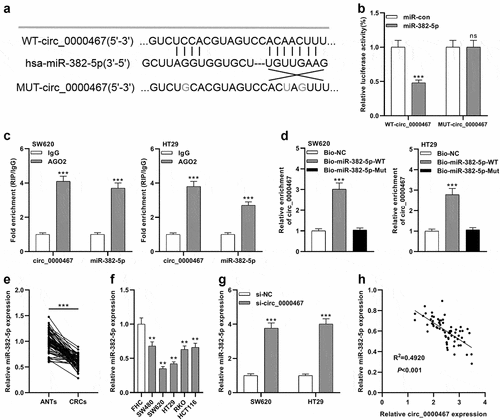
Figure 4. EN2 was a direct target of miR-382-5p
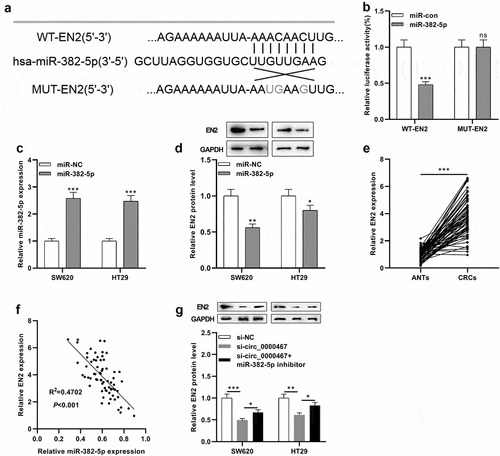
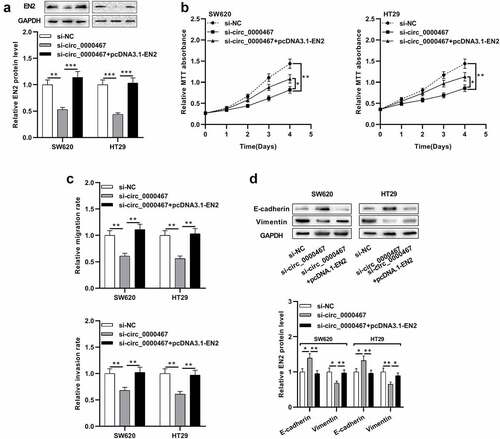
Figure 5. Circ_0000467 enhanced the multiplication and invasion of CRC cells via miR-382-5p/EN2 axis
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.