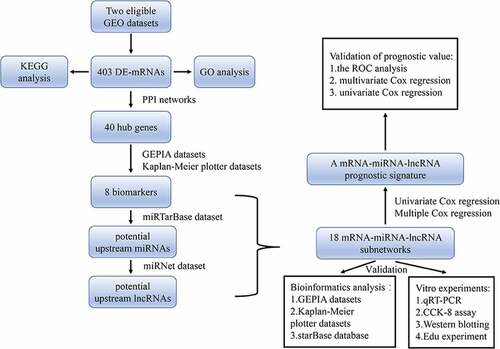
Figures & data
Table 1. The correlations between miRNA-mRNA/lncRNA-mRNA pairs identified by starBase database
Figure 1. Identification of DE-mRNAs in two GEO datasets
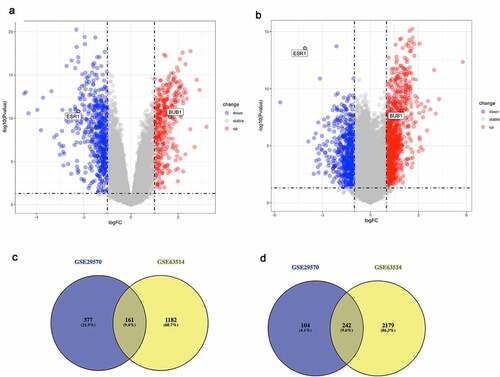
Figure 2. Functional analysis for the DE-mRNAs and identification of hub genes in protein-protein network
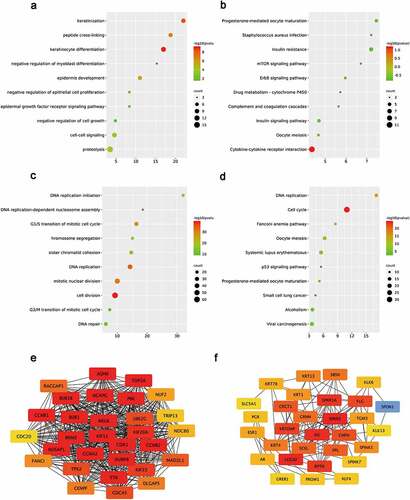
Figure 3. Screening the biomarkers in cervical cancer
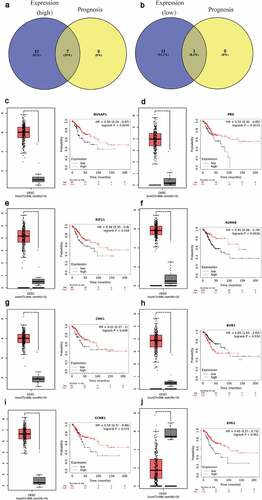
Figure 4. Screening the key lncRNA in cervical cancer
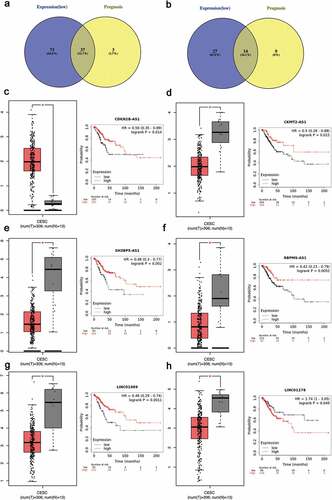
Figure 5. Validation of ce-RNA network
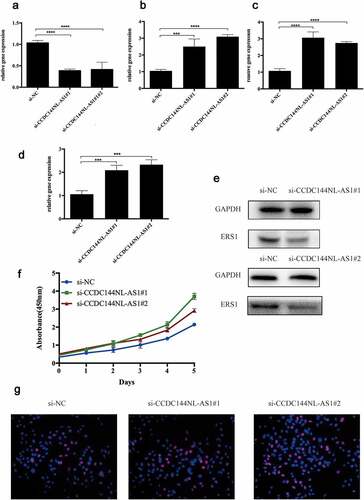
Figure 6. The Cox regression analysis for evaluating the independent prognostic value of the risk score
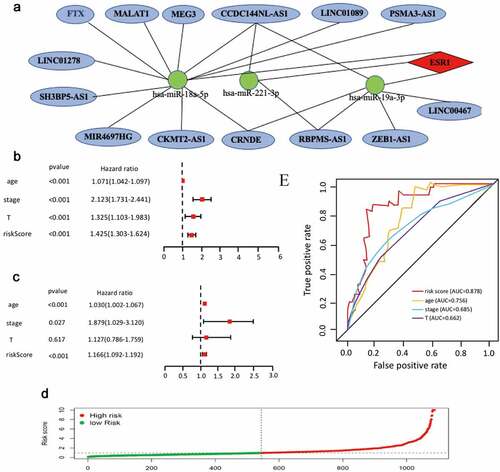
Table 2. The expression levels of these eight RNAs