Figures & data
Table 1. Primers for qRT-PCR
Table 2. The association of miR-1269b expression with clinicopathological parameters
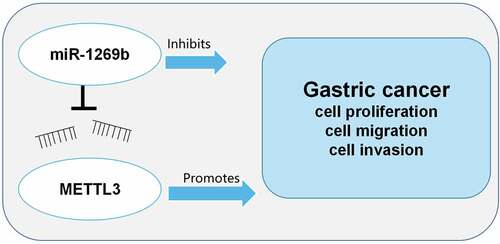
Figure 1. The expression of miR-1269b is inhibited in GC tissues and cells(a) Expression levels of miRNAs in GC tissues (Tumor) and adjacent normal tissues (Normal) in GSE93415. (b) The expression of miR-1269b in GC tissues and adjacent tissues was detected by qRT-PCR. (c) The expression of miR-1269b in GC cells and GES-1 cells was detected by qRT-PCR. (d) Difference in the expression level of miR-1269b between intestinal type GC tissues and diffuse type GC tissues. (e and f) The expression of miR-1269b in GC tissues with different clinical stage (e) and T stage (f). *P < 0.05, **P < 0.01, and ***P < 0.001
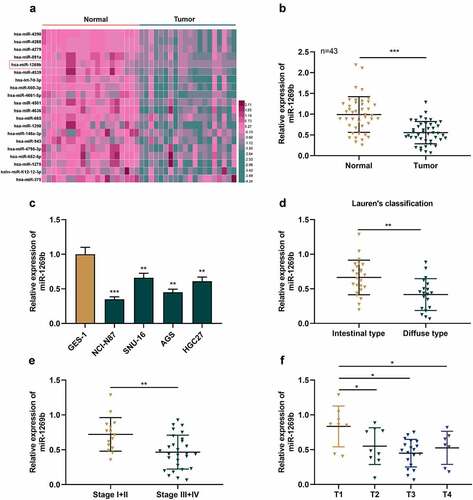
Figure 2. miR-1269b inhibits the proliferation, migration and invasion of GC cells (a) miR-1269b expression in NCI-N87 or SNU-16 cells transfected with miR-1269b mimics or inhibitors were detected by qRT-PCR. (b) CCK-8 assay was used to evaluate cell proliferation. (c) The migration ability of cells was evaluated by wound healing assay. (d) The migration and invasion ability of cells were evaluated by transwell assay. *P < 0.05, **P < 0.01, and ***P < 0.001
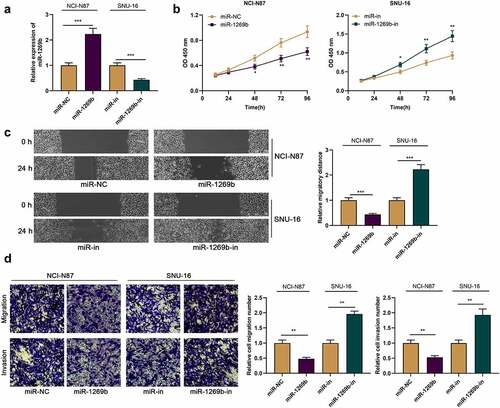
Figure 3. METTL3 is the target of miR-1269b A) The predicted binding sequence between METTL3 mRNA 3ʹUTR and miR-1269b. (b) Dual-luciferase reporter gene assay was used to verify the binding relationship between METLL3 mRNA 3ʹ UTR and miR-1269b. (c) and (d) qRT-PCR and Western blot were used to detect the expression of METTL3 mRNA and protein in NCI-N87 and SNU-16 cells transfected with miR-1269b mimics or inhibitors. (e) qRT-PCR was used to detect the expression of METTL3 mRNA in GC tissues and adjacent tissues. (f) Pearson correlation was used to analyze the correlation between METTL3 mRNA expression and miR-1269b expression in GC tissues. **P < 0.01, and ***P < 0.001
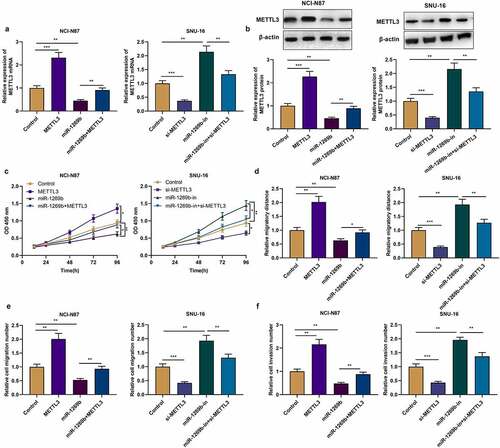
Figure 4. miR-1269b regulates the progression of GC by targeting METTL3 (a) and (b) qRT-PCR and Western blot were used to detect the expression of METTL3 mRNA and protein after the transfection. (c) CCK-8 assay was used to evaluate GC cell proliferation. (d)–(f) The migration and invasion ability of NCI-N87 and SNU-16 cells were evaluated by wound healing assay and transwell assay. *P < 0.05, **P < 0.01, and ***P < 0.001
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.