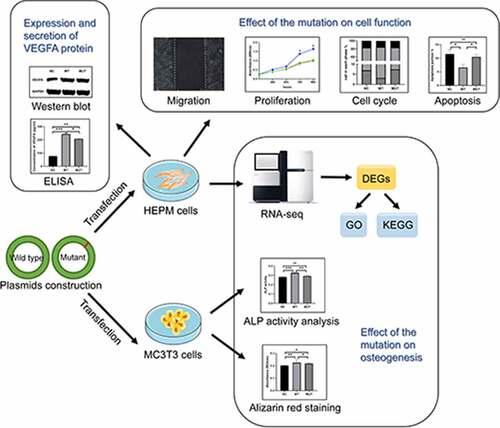
Figures & data
Figure 1. The expression of VEGFA in human cell lines. HEPM and HEK-293 cells were transfected to express either the wild-type/mutant VEGFA allele or control vector. (a) The VEGFA protein expression levels were estimated by western blot (left); the amounts of VEGFA were determined by densitometry of protein bands (right); GAPDH was the internal control. (b) The VEGFA protein levels in cell supernatants were estimated by ELISA. n = 3 for each group. *P < 0.05, **P < 0.01, ***P < 0.001, no significance
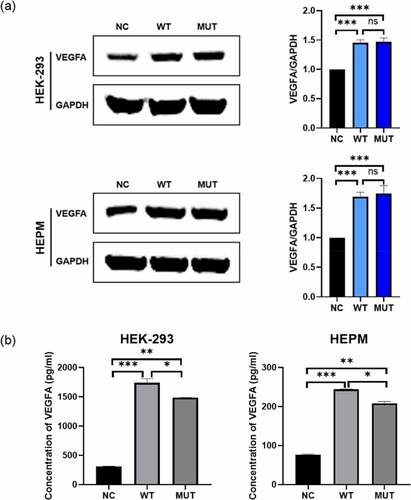
Table 1. qPCR primer sequences
Figure 2. In vitro functional roles of wild-type/mutant VEGFA allele in HEPM and HEK-293 T. (a, b) Effect of VEGFA expression on cell proliferation. (a) HEPM and HEK-293 cells were transfected with wild-type VEGFA, mutant VEGFA or control vector (negative control), cell proliferation was detected every 24 h for 96 h by CCK-8 assay. (b) Cells were seeded in E-Plate L8 plates. After transfection with wild-type VEGFA, mutant VEGFA or control vector, cell proliferation was monitored every 15 minutes by the RTCA iCELLigence instrument. Cell proliferation in wild type group was significantly higher than in mutation group and negative control group. (c) Effects of different type of VEGFA on the cell cycle. Overexpression of wild-type VEGFA alleles decreased the number of cells stagnant G1 phase, but mutant VEGFA alleles did not have the same effect. (d) Effects of VEGFA expression on cell apoptosis. The apoptosis rate was lower in both two kinds of cells overexpressing mutant VEGFA alleles than in the corresponding wild-type VEGFA alleles group. (e, f) The wound healing assay was performed in HEPM (e) and HEK-293 (f) cells. Cells were treated with wild-type/mutant VEGFA plasmid or control vector after scratching. HEPM and HEK-293 T cells overexpressing wild-type VEGFA allele migrated 42% and 32% quicker than cells overexpressing mutant VEGFA allele, respectively. n = 3 for each group. *P < 0.05, **P < 0.01, ***P < 0.001. ns, no significance
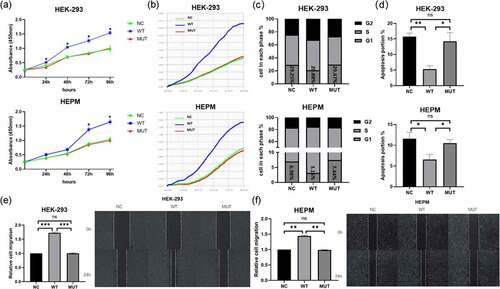
Figure 3. Differential analysis, GO analysis and KEGG pathway analysis of differential gene expression between cells overexpressing wild-type VEGFA and mutant VEGFA. (a) Volcano Plot. Compared with the wild-type VEGFA group, mutant VEGFA triggered 4705 DEGs, containing 2488 downregulated genes (green dot) and 2217 upregulated genes (red dot). (b) GO analysis histogram. 30 enrichment GO entries were shown, according to biological processes, cellular components, and molecular functions. (c) KEGG pathway analysis scatter diagram. 20 significant enriched pathways, including MAPK signaling pathway, TNF signaling pathway, TGFβ signaling pathway and osteoclast differentiation, were shown
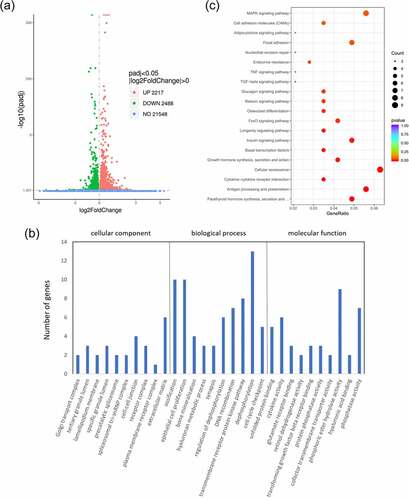
Figure 4. Expression of osteogenesis-related genes by qPCR analysis. The expression of Runx2, OSX, BMP2, ALP, OCN and TGFβ1 was significantly higher in wild type VEGFA group, and had no significant in mutant VEGFA group, compared with the control group. Most importantly, the expression of these genes in the mutant VEGFA group was significantly lower than in the wild type group
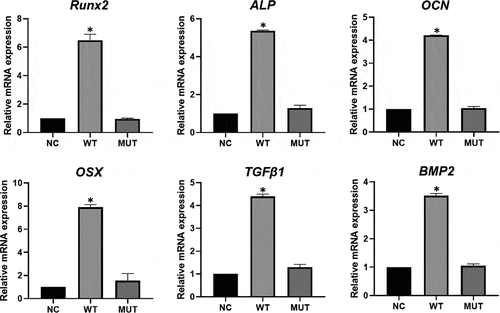
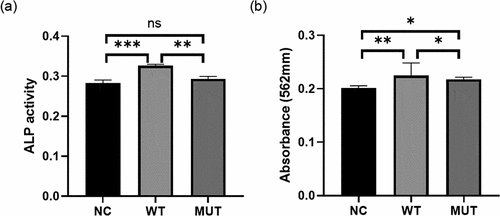
Figure 5. VEGFA promotes osteogenesis, while the mutation of VEGFA reduced the osteogenic effect. (a) ALP activity analysis. MC3T3 cells transfected with wild-type/mutant VEGFA plasmids or control vectors were cultured in osteogenic medium for 3 days and stained by ALP content. (b) Alizarin red stain. The transfected MC3T3 cells cultured for 7 days were stained by alizarin red. n = 3 for each group. *P < 0.05, **P < 0.01, ***P < 0.001. ns, no significance