Figures & data
Table 1. Primer sequences
Table 2. The relationship between circ_0062389 expression and clinicopathological features
Figure 1. Circ_0062389 is significantly up-regulated in PTC tissues and cell lines (a)&(b) Volcanic and heat maps showed that multiple circRNAs, including Circ_0062389, were differentially expressed in PTC tissues and adjacent tissues in GSE93522; (c) qRT-PCR was used to detect the expression of circ_0062389 in PTC tissues and adjacent tissues; (d) qRT-PCR was performed to evaluate the expression of circ_0062389 in PTC cell lines and normal cell line; (e)&(f) After RNase R treatment, qRT-PCR was used to detect the expression of circ_0062389 and PI4KA mRNA in total RNA. ***P < 0.001
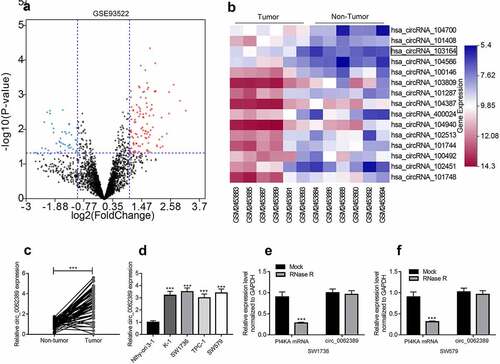
Figure 2. Circ_0062389 can promote the proliferation, migration and EMT process of PTC cells (a)&(b) qRT-PCR was used to detect the expression of circ_0062389 after transfection of three circ_0062389 siRNA into PTC cells; (c)&(d) CCK-8 assay was used to evaluate the proliferation of PTC cells transfected with circ_0062389 siRNA; (e) Wound healing assay was adopted to measure cell migration in PTC cells after transfection of circ_0062389 siRNA; (f)&(g) qRT-PCR and Western blot assay were used to detect the expression of E-cadherin and N-cadherin at mRNA and protein levels in PTC cells transfected with circ_0062389 siRNA. **P < 0.01, and ***P < 0.001
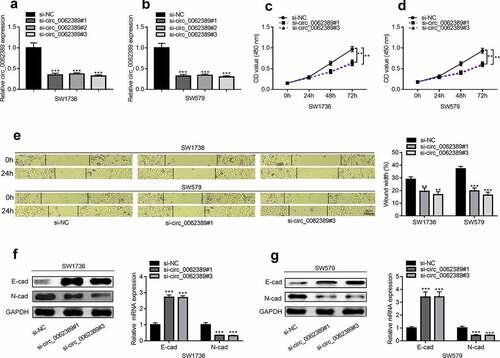
Figure 3. MiR-1179 is a target of circ_0062389 (a)StarBase database predicted that there was a binding site between miR-1179 and circ_0062389; (b)&(c) Dual-luciferase reporter gene assay confirmed that miR-1179 could negatively regulate the luciferase activity of circ_0062389-WT, rather than that of circ_0062389-MUT; (d) qRT-PCR was used to detect miR-1179 expression after PTC cells were transfected with circ_0062389 siRNA; (e) StarBase database was adopted to analyze miR-1179 expression in PTC tissue and normal tissue; (f) qRT-PCR was used to detect the expression of miR-1179 in PTC tissues and adjacent tissues; (g) qRT-PCR was used to detect miR-1179 expression in PTC cell lines and normal cell line; (h) The correlation analysis of circ_0062389 and miR-1179 expression in PTC tissues. **P < 0.01; and ***P < 0.001
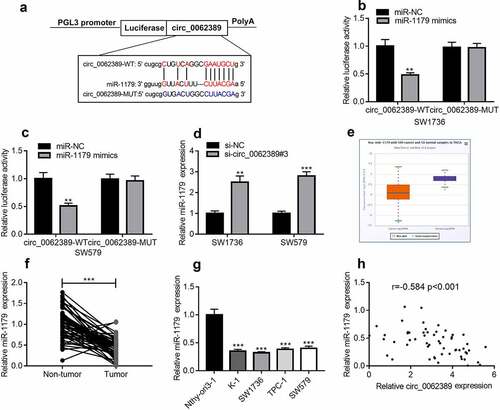
Figure 4. Circ_0062389 exerts promoting effects in PTC through modulating miR-1179. (a) qRT-PCR was used to detect the expression of miR-1179 after transfection of miR-1179 inhibitors into PTC cells; (b)&(c) CCK-8 assay was used to detect the proliferation of PTC cells after co-transfection of circ_0062389 siRNA and miR-1179 inhibitors; (d) Wound healing assay was used to detect PTC cell migration after co-transfection of circ_0062389 siRNA and miR-1179 inhibitors into PTC cells; (e)&(f) qRT-PCR was used to detect the mRNA expression of E-cadherin and N-cadherin after co-transfection of circ_0062389 siRNA and miR-1179 inhibitors into PTC cells. *P < 0.05, **P < 0.01, and ***P < 0.001
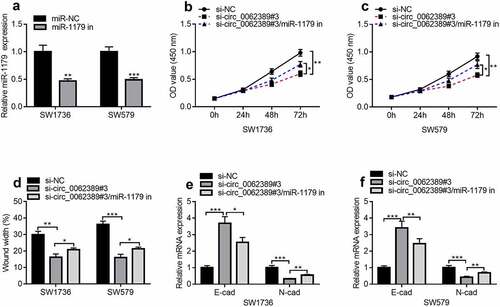
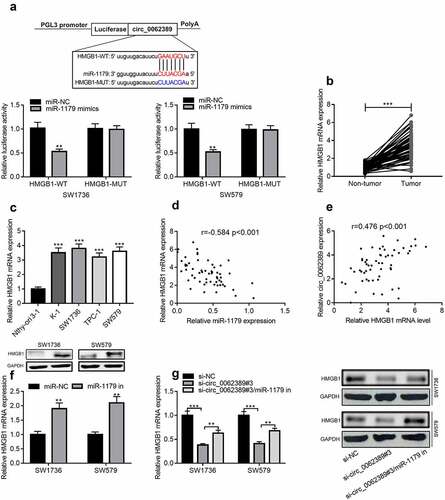
Figure 5. Circ_0062389 can adsorb miR-1179 to regulate HMGB1 expression (a) StarBase database predicted that circ_0062389 and miR-1179 had a complementary binding site, and dual-luciferase reporter gene asasy confirmed that circ_0062389 could bind with miR-1179 directly; (b) qRT-PCR was used to evaluate the expression of HMGB1 mRNA in PTC tissues and adjacent tissues; (c) qRT-PCR was used to detect HMGB1 mRNA expression in PTC cell lines and human normal thyroid follicular epithelial cell line; (d)&(e) The correlation analysis of miR-1179 and HMGB1 mRNA, HMGB1 mRNA and circ_0062389 in PTC tumor tissues; (f)&(g) qRT-PCR and Western blot assay were used to detect the expression of HMGB1 mRNA and protein in PTC cells after transfection of miR-1179 inhibitors and co-transfection of circ_0062389 siRNA and miR-1179 inhibitors. **P < 0.01, and***P < 0.001
Supplemental Material
Download ()Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.