Figures & data
Table 1. The PCR primer sequence used in this study
Table 2. Chi-square test of RB patient characteristics and XIST expression
Figure 1. The expression of XIST in RB tissues and cell lines. (a) The expression of XIST in normal and RB tissues was detected by qRT-PCR. (b) The expression of XIST in RB cell lines and normal human retinal pigment epithelial cells was detected by qRT-PCR
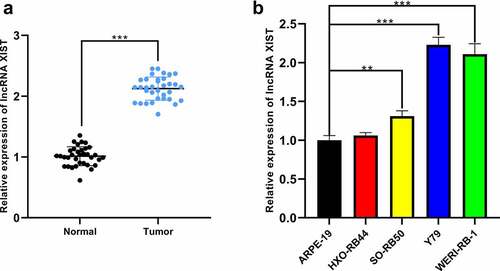
Figure 2. The biological effects of XIST on the malignant phenotypes of RB cells. (a-b) Cell models of XIST overexpression and knockdown were established in SO-Rb50 and Y79 cell lines, respectively. (c-d) The effect of XIST overexpression or knockdown on RB cell proliferation was detected by CCK-8 assay. (e-f) The effect of XIST overexpression or knockdown on RB cell migration and invasion was detected through Transwell experiment. (g) The effect of XIST overexpression or knockdown on RB cell apoptosis was assessed through TUNEL experiment
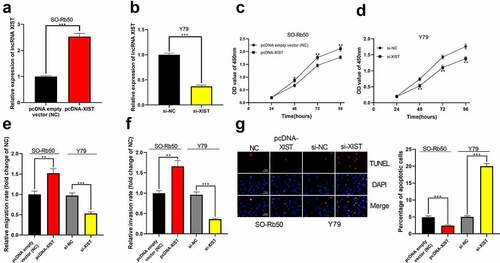
Figure 3. The interaction between miR-191-5p and XIST. (a) Multiple miRNAs including miR-191-5p were abnormally expressed in GSE7072. (b) The binding site between XIST and miR-191-5p was predicted by StarBase database. (c) The expression of miR-191-5p in normal tissues and RB tissues was detected by qRT-PCR. (d) The expression of miR-191-5p in RB tissue was negatively correlated with XIST in RB samples. (e) The expression of miR-191-5p in RB cell lines and normal human retinal pigment epithelial cell was detected by qRT-PCR. (f-g) The targeted binding relationship between XIST and miR-191-5p was verified through the dual-luciferase reporter gene assay
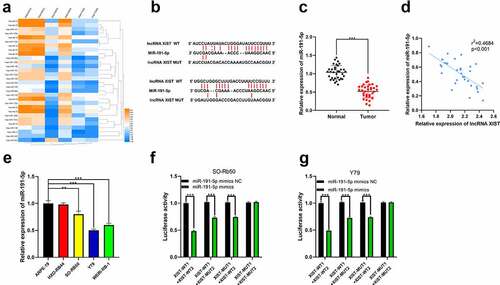
Figure 4. The effect of miR-191-5p on RB cells. (a-b) MiR-191-5p overexpression and inhibition cell models were established in Y79 and SO-Rb50 cells, respectively. (c-d) Through CCK-8 assay, the effect of miR-191-5p overexpression or inhibition on RB cell proliferation was detected. (e-f) The effect of miR-191-5p overexpression or inhibition on RB cell migration and invasion was detected by transwell experiment. (g) The effect of miR-191-5p overexpression or inhibition on RB cell apoptosis was detected by TUNEL experiment
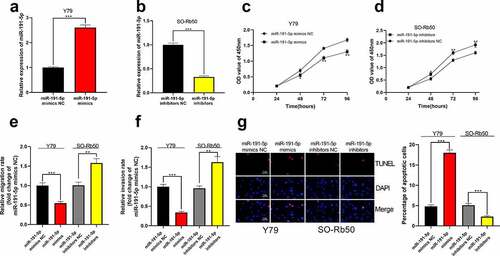
Figure 5. The relationship between miR-191-5p and BDNF. (a) The target genes of miR-191-5p were predicted through microT, miRanda, miRmap, and target scan databases. (b) The predicted binding site between miR-191-5p and BDNF 3ʹUTR. (c) The expression of BDNF in normal tissues and RB tissues was detected by qRT-PCR. (d) The expression of BDNF in RB was negatively correlated with miR-191-5p expression in RB samples. (e) The expression of BDNF in RB cell line was detected by Western blot. (f-g) The targeted binding relationship between miR-191-5p and BDNF was verified through a dual-luciferase reporter gene assay. (h) The expression levels of BDNF mRNA in miR-191-5p overexpression and inhibition models were detected by qRT-PCR. (i) Western blot was used to detect the expression level of BDNF in miR-191-5p overexpression and inhibition models
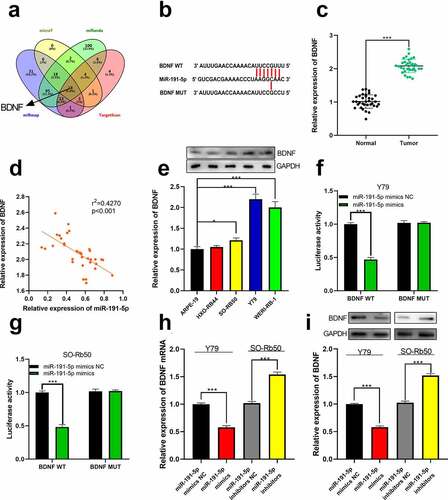
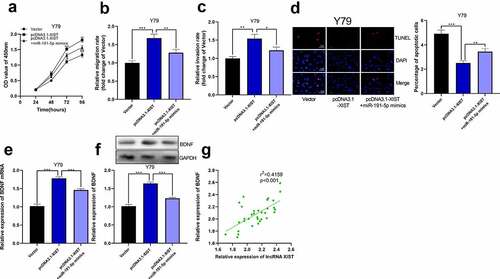
Figure 6. The overexpression of miR-191-5p reversed the effect of XIST on RB cells. (a) CCK-8 assay was used to detect the effect of co-transfected miR-191-5p mimics on the proliferation of RB cells with XIST overexpression. (b-c) The effect of co-transfected miR-191-5p mimics on the migration and invasion of RB cells with XIST overexpression was detected by transwell experiment. (d) The effect of co-transfected miR-191-5p mimics on apoptosis of RB cells with XIST overexpression was detected by TUNEL experiment. (e) The expression levels of BDNF mRNA in control, pcDNA3.1-XIST, and pcDNA3.1-XIST+miR-191-5p mimics groups were detected by qRT-PCR. (f) The expression levels of BDNF in control, pcDNA3.1-XIST, and pcDNA3.1-XIST+miR-191-5p mimics groups were detected by Western blot. (g) The expression of BDNF in RB tissue was positively correlated with the expression of XIST