Figures & data
Figure 1. Construction of WGCNA analysis
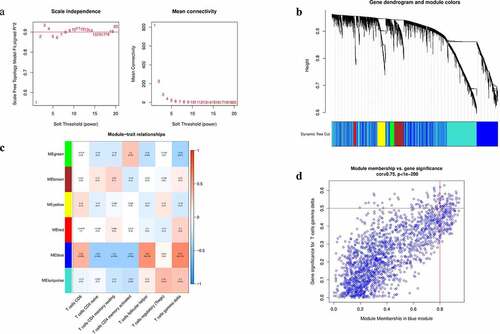
Figure 2. The enrichment analysis of hub module
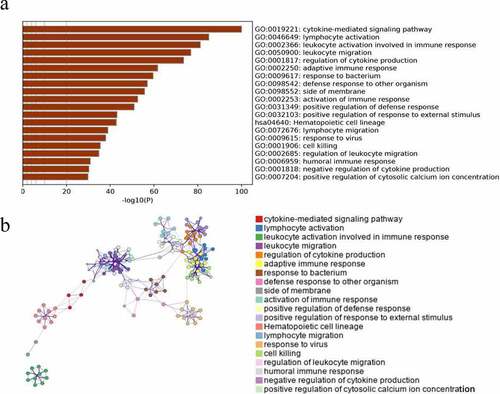
Figure 3. Identification of hub genes
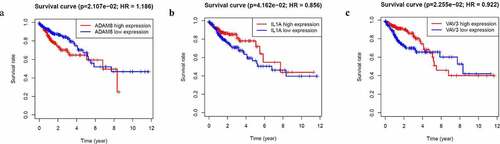
Figure 4. Correlation between three expressed prognostic genes and immune cell infiltration through TIMER
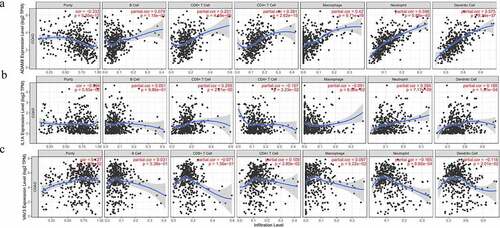
Figure 5. Genetic Alteration of hub genes in CRC

Figure 6. Co-expression Genes correlated with hub genes in CRC
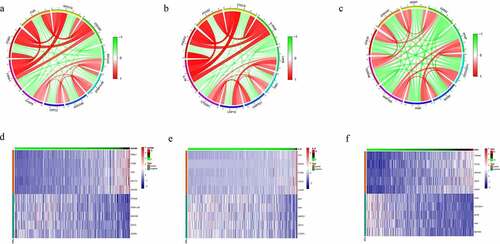
Figure 7. Function analysis and signaling pathways of hub genes
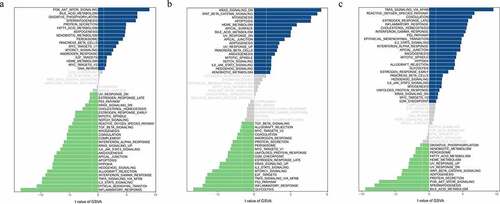
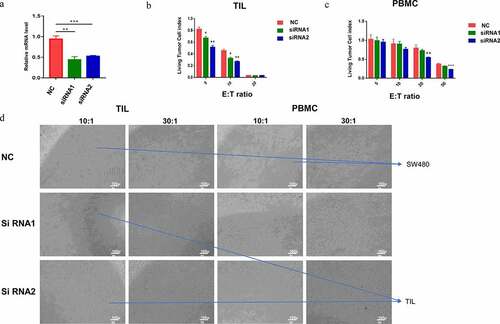
Figure 8. Functional experiments of ADAM8
Supplemental Material
Download ()Data availability statement
Data subject to third-party restrictions. The data were obtained from TCGA(https://tcga-data.nci.nih.gov/tcga/).