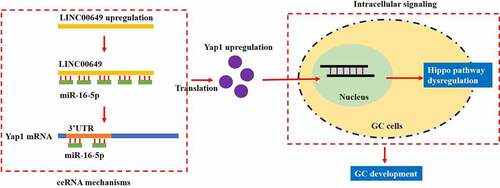
Figures & data
Figure 1. LINC00649, miR-16-5p and YAP1 were relevant to GC malignancy. The expression status of (a) LINC00649, (b) miR-16-5p, and (c) YAP1 mRNA in the clinical specimens were quantified by using the Real-Time qPCR analysis. (d-f) The correlations of LINC00649, miR-16-5p and YAP1 mRNA in the GC tissues were analyzed by pearson correlation analysis. (g-i) Real-Time qPCR was used to detect LINC00649, miR-16-5p and YAP1 mRNA expression levels in GC cells, and (j, k) the YAP1 protein levels in the cells were measured by using the Western Blot analysis. Each experiment had three individual repetitions, and *P < 0.05 was regarded as statistical significance
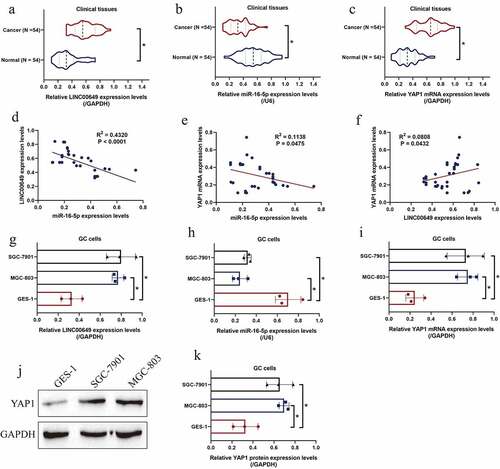
Figure 2. Targeting the LINC00649 hindered cell proliferation, viability, EMT and tumorigenesis in GC. (a, b) The transfection efficiency for LINC00649 overexpression and downregulation vectors were determined by Real-Time qPCR analysis. (c, d) MTT assay was performed to examine cell proliferation abilities. (e) Cell viability was determined by trypan blue staining assay. (f) In vivo tumors were obtained and weighed to evaluate tumorigenesis of MGC-803 cells. (g) Transwell assay was performed to determine cell migration abilities. (h-k) The EMT-associated biomarkers in GC cells were examined by using the Western Blot analysis. (l) The GC cells were stained with Annexin V-FITC and PI to examine cell apoptosis ratio. Each experiment had three individual repetitions, and *P < 0.05 was regarded as statistical significance
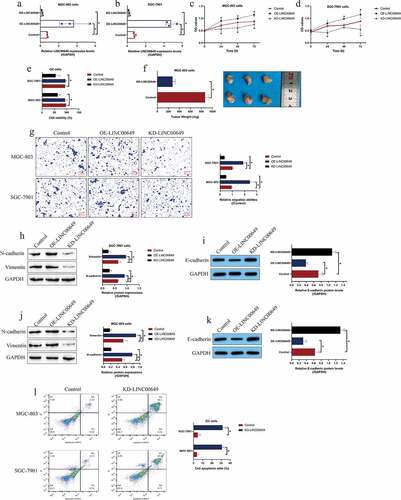
Figure 3. MiR-16-5p was able to target both LINC00649 and YAP1 in GC cells. (a, d) The tarting sites among LINC00649, miR-16-5p and YAP1 were predicted by miRDB software, and (B-C, E-F) the above targeting sites were validated by performing the following dual-luciferase reporter gene system assay. (g-j) RNA pull-down assay was performed to evaluate the binding abilities of miR-16-5p with LINC00649 and YAP1. (k, l) Real-Time qPCR and (m, n) Western Blot validated that LINC00649 positively regulated YAP1 via sponging miR-16-5p. (o, p) The mRNA levels of EGFR, SOX2 and OCT4 were examined by using the Real-Time qPCR analysis. Each experiment had three individual repetitions, and *P < 0.05 was regarded as statistical significance
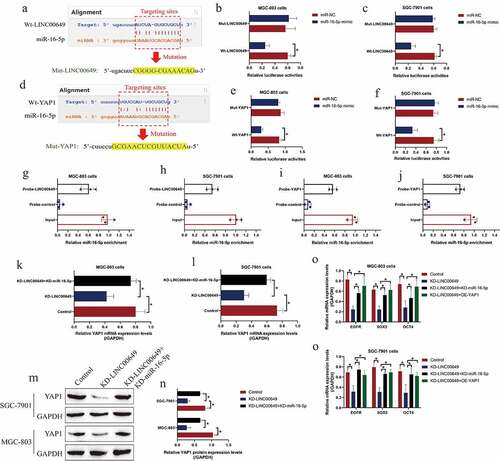
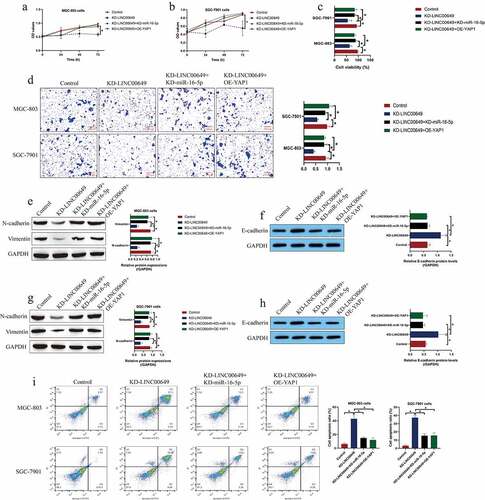
Figure 4. Silencing of LINC00649 reversed the malignant phenotypes in GC cells by regulating the miR-16-5p/YAP1 axis. (a, b) MTT assay was performed to evaluate cell proliferation abilities. (c) Trypan blue staining assay was performed to detect cell viability. (d) Cell migration abilities were evaluated by using the Transwell assay. (e-h) The expression levels of N-cadherin, Vimentin and E-cadherin were examined by Western Blot analysis. (i) FCM assay was used to evaluate cell apoptosis ratio in the GC cells. Each experiment had three individual repetitions, and *P < 0.05 was regarded as statistical significance