Figures & data
Table 1. Clinical and pathological characteristics of bladder cancer samples in this study
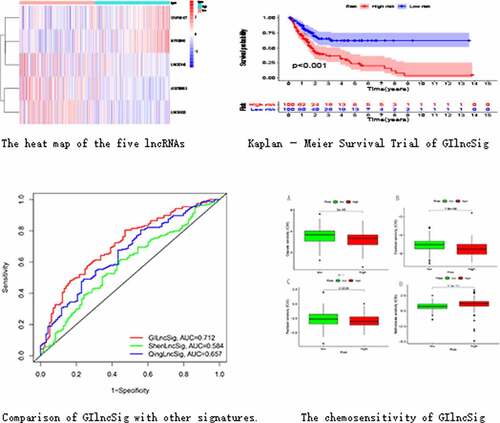
Figure 1. Heatmap of expression of 58 differential lncRNAs. The abscissa is the bladder cancer sample, which is divided into GS-like group (blue) and GU-like group (red), and the ordinate is 58 lncRNAs
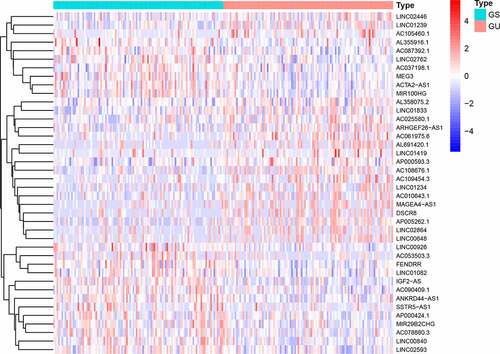
Figure 2. Co-expression network and enrichment analysis of differential lncRNAs. (a) Expression networks of differential lncRNAs and their associated mRNAs. The blue circles represent lncRNAs and red circles represent mRNAs. (b) Gene Ontology (GO) enrichment analysis and (c) Kyoto Encyclopedia of Gene and Genomes (KEGG) pathway analysis of mRNAs associated with differential lncRNAs
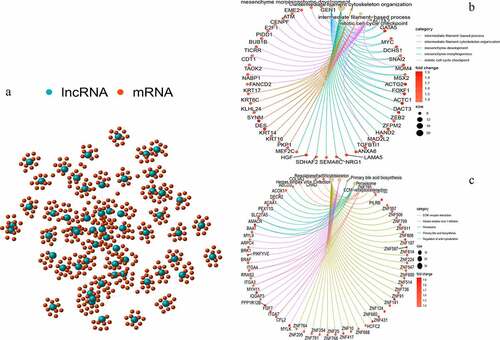
Table 2. LncRNAs associated with the prognosis of bladder cancer patients obtained after multivariate COX analysis
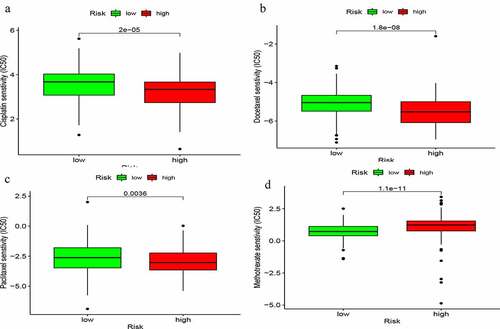
Figure 3. Identification of genomic instability-related lncRNA signature (GIlncSig). (a) Prognostic relevant lncRNAs obtained after univariate COX analysis. (b) Expression heatmap of five lncRNAs obtained after multivariate COX analysis. The abscissa is the bladder cancer sample, with blue representing the high-risk group and red representing the low-risk group
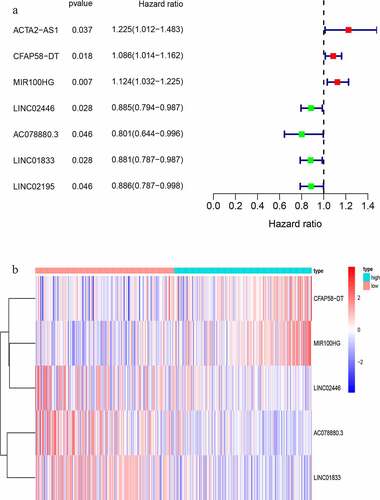
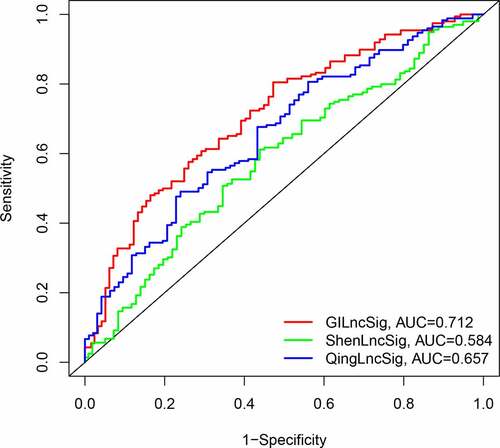
Figure 4. Evaluation of instability-related lncRNA genomic signature (GIlncSig) in the training group. (a) Kaplan – Meier Survival Trial of GIlncSig for High and Low Risk Groups. (b) Risk score for each tumor sample. (c) Survival outcome for each tumor sample. (d) Time-dependent ROC curve at 5 years
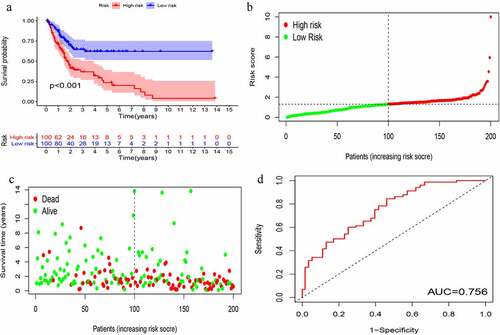
Figure 5. Evaluation of instability-related lncRNA genomic signature (GIlncSig) in the testing group. (a) Kaplan – Meier Survival Trial of GIlncSig for High and Low Risk Groups. (b) Risk score and (c) Survival outcome for each tumor sample. (d) Time-dependent ROC curve at 5 years
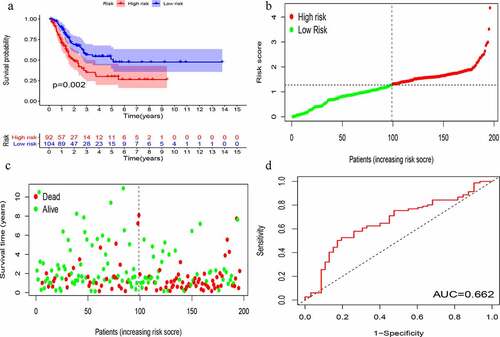
Table 3. Independence of GIlncSig by univariate and multivariate COX analysis
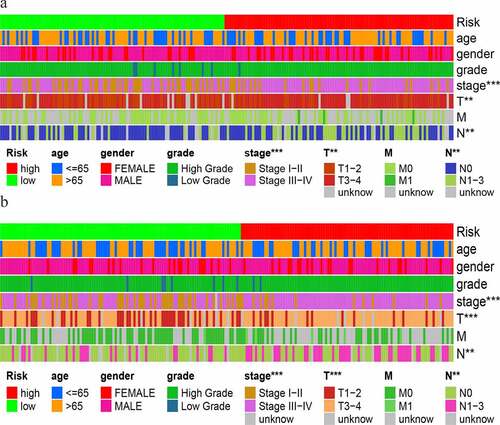
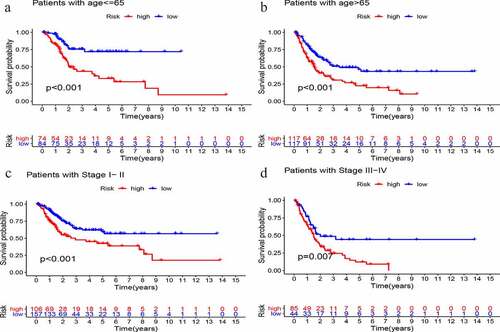
Figure 7. Stratified analysis of GIlncSig. Survival curves for high and low risk groups in patients ≤ 65 years (a) and > 65 years (b). Survival curves for high and low risk groups in early stage (c) and advanced stage patients (d)
Supplemental Material
Download ()Dat availability statement
All the data sets of this study are available in the TCGA database(https://portal.gdc.cancer.gov/) and GEO database (http://www.ncbi.nlm.nih.gov/geo/).