Figures & data
Figure 1. Circ_0008274 expression is significantly up-regulated in HCC tissues and cells
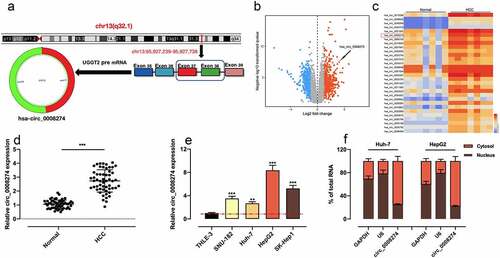
Table 1. Primers used for qRT-PCR
Table 2. Correlation between clinicopathological features and expression of circ_0008274 in HCC tissues
Figure 2. Circ_0008274 promotes the proliferation, migration and invasion of HCC cells
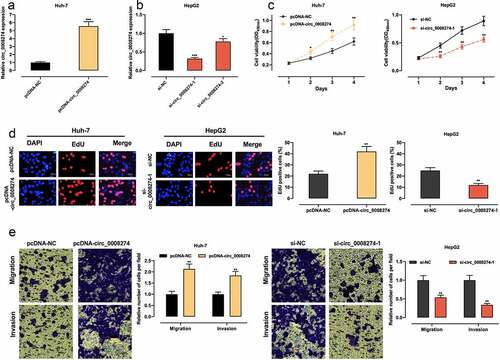
Figure 3. Circ_0008274 adsorbs miR-140-3p
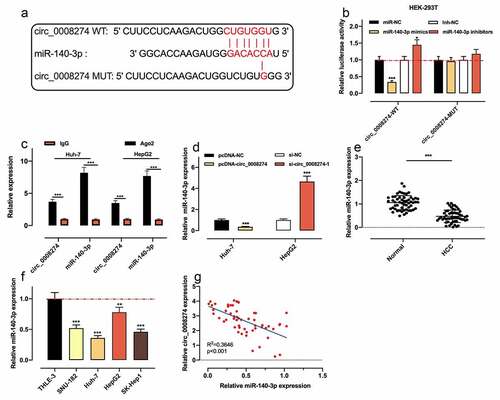
Figure 4. Circ_0008274 can promote the malignant behaviors of HCC cells by targeting miR-140-3p
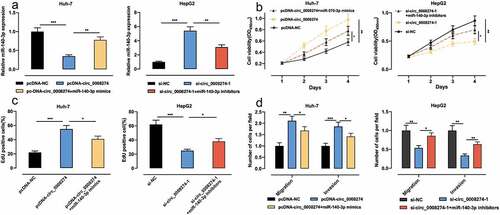
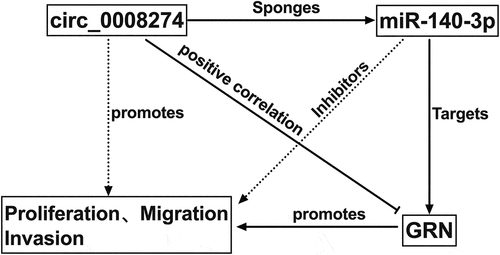
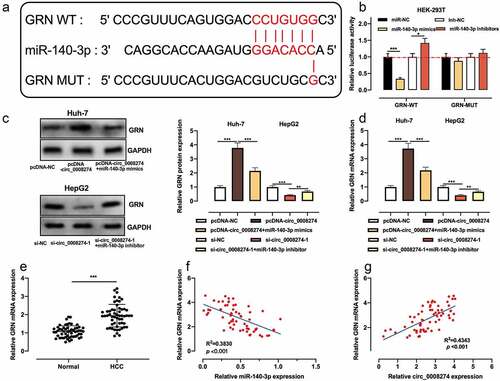
Figure 5. Circ_0008274 regulates miR-140-3p/GRN axis
Supplemental Material
Download ()Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.