Figures & data
Figure 1. A) Q-Q Plot of C50-C50 (β = 1.05, λmean = 1.07, λmedian = 1.06) Containing 9,113,133 imputed variants that passed quality control (QC), with a P different than 0; b) Manhattan PLOT (IMPUTED) CONTAINING all (QC and non-QC) 30,798,054 imputed variants; c) Venn diagram of significant genes (CLDN7,MLLT10,RBM33,SH3RF1,SSBP4, UBE2Z,BMPER,FGF7,MSRB3,TNRC6B) in breast cancer (BC)
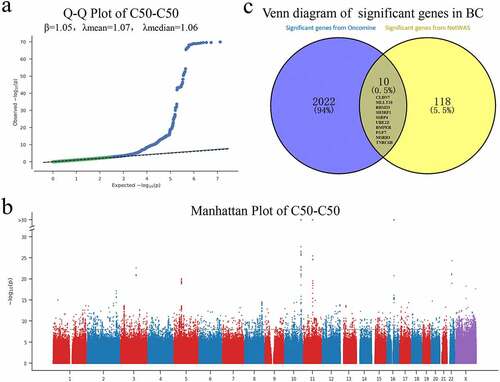
Figure 2. Heatmap of significant different expression genes in BC. a)Top 20 Over-expressed genes; b) Top 20 Under-expressed genes
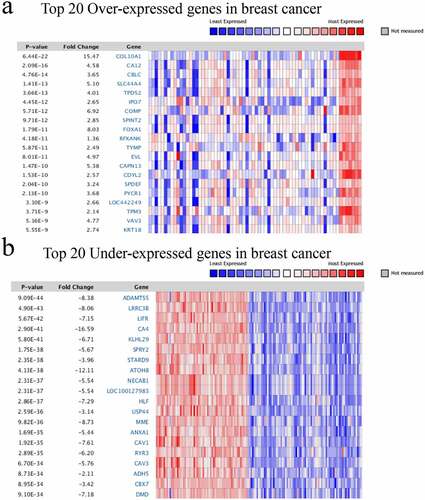
Table 1. Significant genes in breast cancer identified using NetWAS
Figure 3 KEGG pathway analysis of significant genes (CLDN7, MLLT10, RBM33, SH3RF1, SSBP4, UBE2Z, BMPER, FGF7, MSRB3 and TNRC6B) in BC
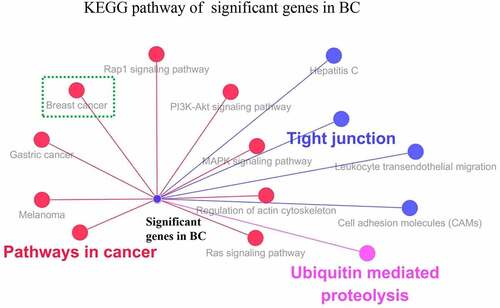
Table 2. Gene ontology (GO) enrichment result of significant genes in breast cancer
Figure 4. The immunohistochemistry of significant genes(CLDN7, RBM33, SH3RF1, UBE2Z, FGF7 and TNRC6B) in BC
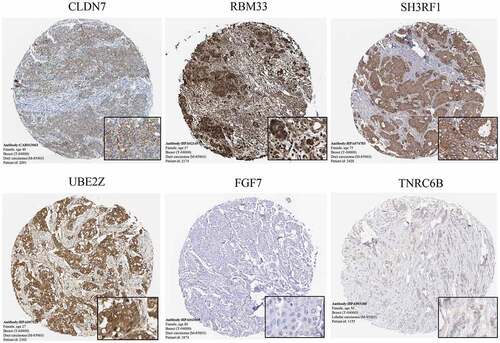
Figure 5. The different expression of significant genes genes(CLDN7, BMPER, FGF7 and MSRB3) in breast cancer samples to normal samples, the red box mean in breast cancer samples and the black box mean in normal samples. *:p < 0.05
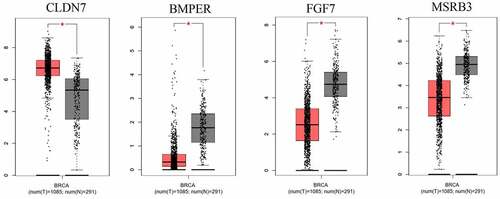
Figure 6. Overall survival analysis combined multiple genes expression. a)Combined significant Over-expressed genes(CLDN7, MLLT10, RBM33, SH3RF1, SSBP4 and UBE2Z) in BC based on GSE58812; b)Combined significant Over-expressed genes(CLDN7, MLLT10, RBM33, SH3RF1, SSBP4 and UBE2Z) in BC based on GSE42568; c)Combined significant under-expressed genes(BMPER, FGF7, MSRB3 and TNRC6B) in BC based on GSE42568; d)Combined BMPER,FGF7,MSRB3 and TNRC6B in BC based on GSE37751; e)Combined BMPER,MSRB3 and TNRC6B in BC based on GSE1456_U133B; f)Combined BMPER,MSRB3 and TNRC6B in BC based on GSE3494_U133B; g)Combined FGF7 and TNRC6B in BC based on GSE3494_U133A; h)Combined FGF7 in BC based on GSE9893
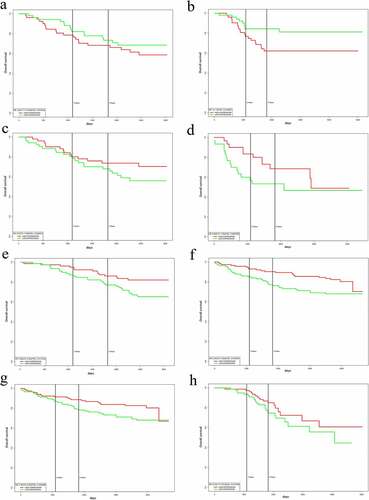
Table 3. The most significant drugs provided by cmap to reverse core genes of breast cancers