Figures & data
Figure 1. Circ_0001162 was highly expressed in glioma. (a–b) The detection of circ_0001162 by qRT-PCR was performed in glioma samples (a) and cells (b). (c–f) Circ_0001162 stability was determined via qRT-PCR following the treatment of Actinomycin D (c–d) or RNase R (e–f). (g–h) The analysis of circ_0001162 was implemented via qRT-PCR using the reverse transcription products with random or oligo (dt)18 primers. (i–j) Circ_0001162 localization was examined via qRT-PCR in LN18 and A172 cells. *P < 0.05
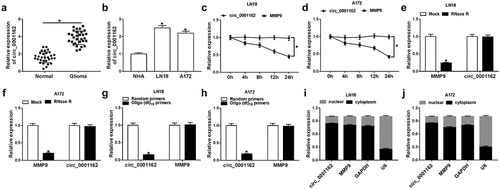
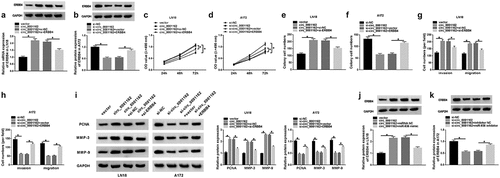
Figure 2. Circ_0001162 facilitated the tumorigenesis of glioma in vitro and in vivo. (a–b) The qRT-PCR analysis of circ_0001162 was performed in LN18 and A172 cells transfected with circ_0001162, si-circ_001162 or their controls. (c–g) The effect of circ_0001162 overexpression (in LN18 cells) or knockdown (in A172 cells) on glioma progression was researched by cell proliferation using MTT (c–d), colony formation by colony formation assay (e) and migration/invasion via transwell assay (f–g). (h) The protein levels of PCNA, MMP-3 and MMP-9 were detected using WB. (i–n) Tumor volume, weight and circ_0001162 expression of glioma were measured after circ_0001162 up-regulation (i–k) or knockdown (l–n). *P < 0.05
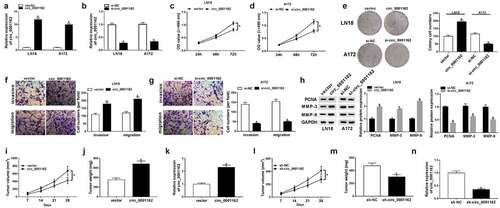
Figure 3. Circ_0001162 directly targeted miR-936. (a) CircRNA interactome was applied for miRNA target prediction of circ_0001162. (b–d) The target relation between circ_0001162 and miR-936 was verified via the dual-luciferase reporter assay. (e) The efficiency of circ_0001162 probe was evaluated via qRT-PCR. (f) The miR-936 level was measured through qRT-PCR in RNA pull-down assay with biotinylated circRNA. (g–h) The detection of miR-936 in glioma tissues and cells was carried out with qRT-PCR. (i–j) The transfection efficiencies of miR-936 mimic (in LN18 cells) and miR-936 inhibitor (in A172 cells) were estimated by qRT-PCR. (k–l) The qRT-PCR was implemented for miR-936 detection in LN18 cells transfected with circ_0001162, circ_0001162+ miR-936 mimic or matched controls and A172 cells with transfection of si-circ_0001162, si-circ_0001162+ miR-936 inhibitor or relative controls. *P < 0.05
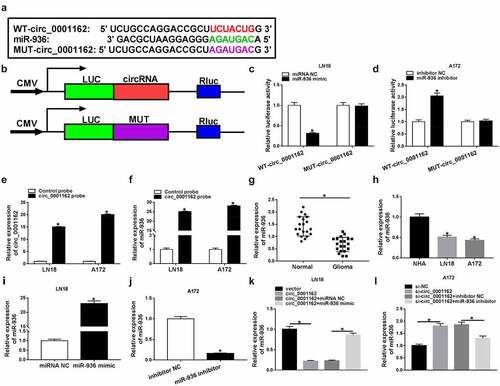
Figure 4. The oncogenic role of circ_0001162 in glioma cells was owed to miR-936. Transfection ofcirc_0001162, circ_0001162+ miR-936 mimic in LN18 cells and si-circ_0001162, si-circ_0001162+ miR-936 inhibitor in A172 cells was performed, including their respective controls. (a–b) The proliferation evaluation was carried out through MTT assay. (c–d) Colony formation analysis was administrated using colony formation assay. (e–f) The determination of cell migration and invasion was conducted via transwell assay. (g) WB was used for the expression analysis of PCNA, MMP-3 and MMP-9. *P < 0.05
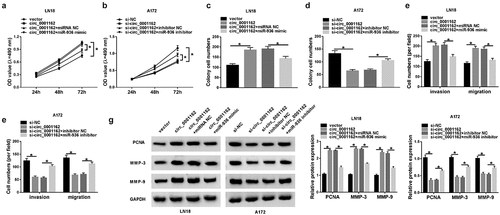
Figure 5. MiR-936 acted as a tumor repressor of glioma via targeting ERBB4. (a) Targetscan exhibited the binding of ERBB4 3ʹUTR and miR-936. (b–c) Whether ERBB4 could bind to miR-936 was verified using dual-luciferase reporter assay. (d–f) WB was administrated to assay the ERBB4 protein level in glioma tissues (d), LN18/A172 cells (e) and the transfection efficiency of ERBB4 and si-ERBB4 (f). (g–h) ERBB4 protein expression was detected via WB after transfection of miR-936 mimic, miR-936 mimic+EREBB4 or controls in LN18 cells (g) and miR-936 inhibitor, miR-936 inhibitor+si-ERBB4 or controls in A172 cells (h). (i–n) Cellular proliferation (i–j), colony formation (k–l) and migration or invasion (m–n) were severally evaluated using MTT, colony formation assay and transwell assay. (o) PCNA, MMP-3 and MMP-9 protein expression detection was performed by WB. *P < 0.05
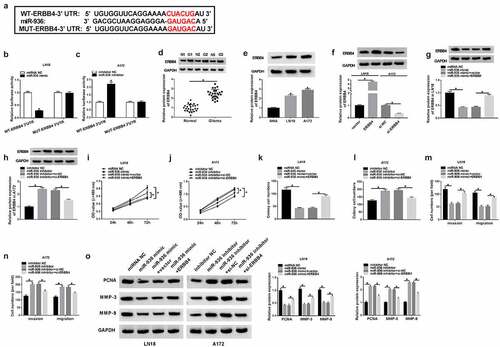
Figure 6. Circ_0001162 contributed to the glioma progression through the regulation of miR-936/ERBB4. (a–b) ERBB4 protein quantification by WB was carried out in LN18 cells with transfection of circ_0001162, circ_0001162+ si-ERBB4 or their controls (a) and A172 cells with transfection of si-circ_0001162, si-circ_0001162+ ERBB4 or respective controls (b). (c–f) The examination of cell proliferation using MTT (c–d) and colony formation via colony formation assay (e–f) was executed in the above groups. (g–h) Transwell assay was adopted for assessing migration and invasion. (i) The measurement of PCNA, MMP-3 and MMP-9 protein expression was completed via WB. (j–k) WB was exploited for estimating the effect of the circ_0001162/miR-936 axis on the ERBB4 protein level. *P < 0.05