Figures & data
Table 1. Characteristics of the study subjects
Figure 1. The alpha diversity and beta diversity indices of the oral microbiota in migraine and control groups
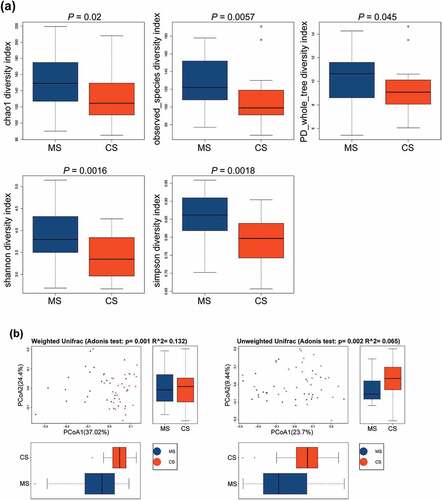
Figure 2. Relative abundances of the oral microbiota in migraine and control groups
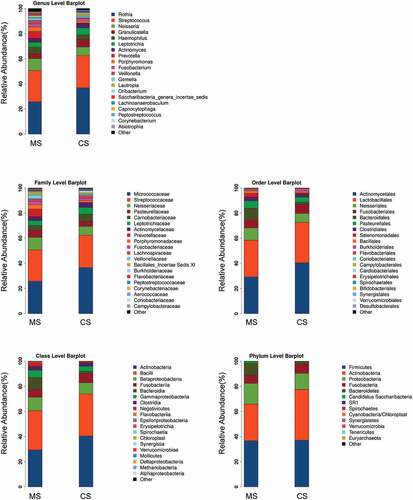
Figure 3. Taxonomic differences of the oral microbiota in migraine and control groups
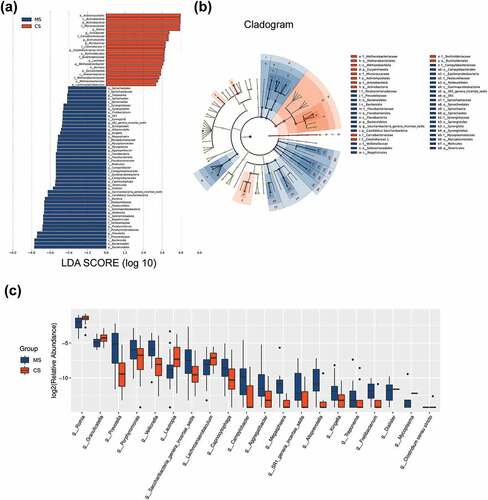
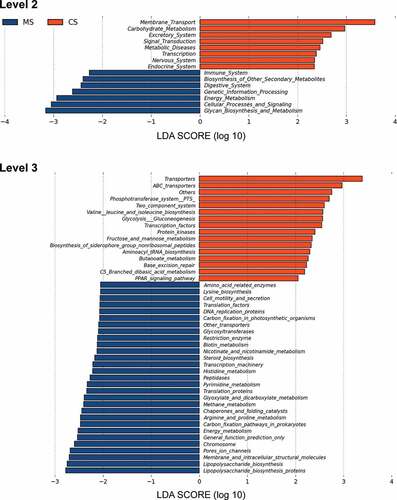
Figure 4. Functional predictions for the oral microbiota of migraine and control groups
Supplemental Material
Download ()Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the [NCBI Sequence Read Archive] repository at [https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA680860].