Figures & data
Table 1. Active compounds and corresponding potential therapeutic targets of YSNJF
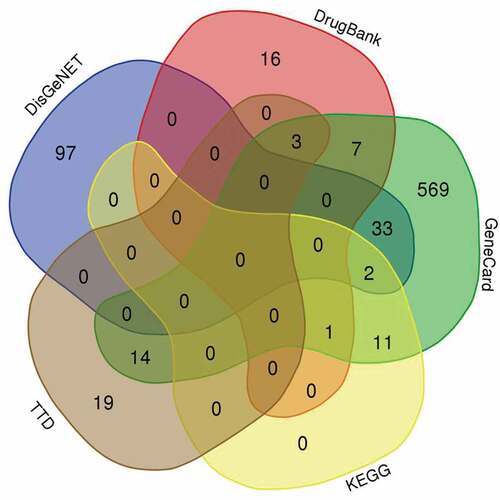
Table 2. Intersected compound from multiple herbal medicines
Table 3. List of major compounds corresponding to intersected targets
Figure 3. YSNJF-compounds-target-MDS network. Blue nodes represent drug and disease, respectively; red nodes represent active compounds; green nodes represent disease-related targets. Lines in the figure represent the interaction between two nodes
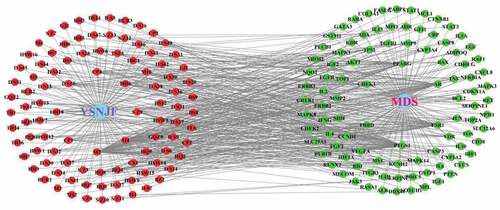
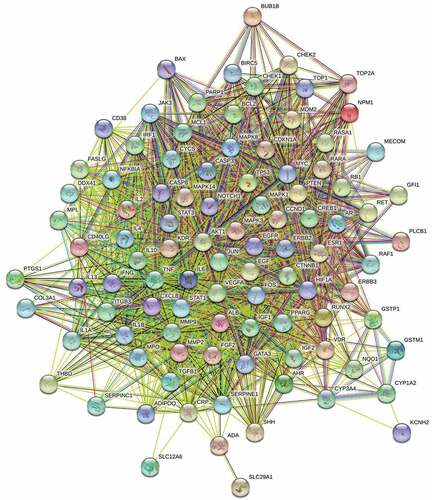
Figure 5. Protein–protein interaction (PPI) network of identified major targets. Light blue nodes were regular targets while yellow nodes were major targets
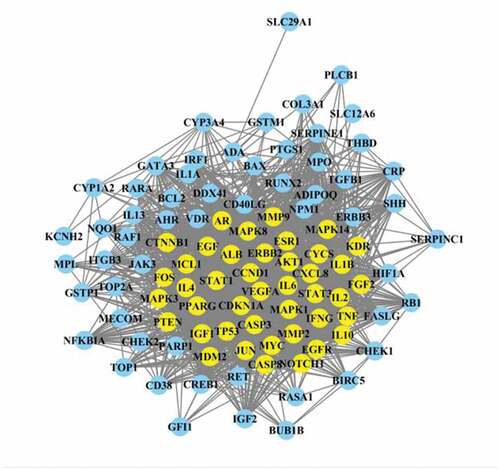
Figure 6. Protein–protein interaction (PPI) network of identified core targets. Light blue nodes were regular targets while yellow nodes were core targets
Figure 7. GO enrichment analysis of intersected targets between drug and disease. The top 20 GO terms in biological process (BP, A), cellular component (CC, B), molecular function (MF, C) with adjusted P value <0.05 were selected and present in a bubble chart manner. The size of bubble represents number of targets enriched in the indicated pathway and the color of the bubble represents the P value of enrichment
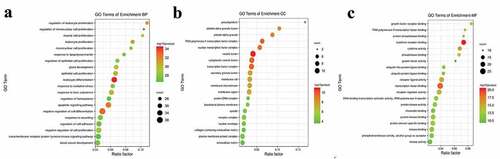
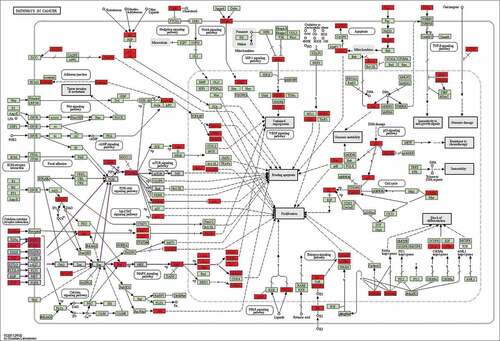
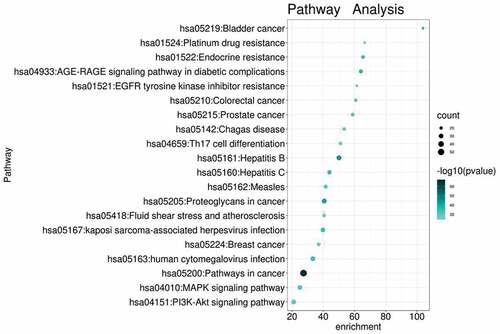
Figure 8. KEGG enrichment analysis of intersected targets between drug and disease. The top 20 KEGG pathways with adjusted P value <0.05 were selected and present in a bubble chart manner. The size of bubble represents the number of targets enriched in the indicated pathway and the color of the bubble represents the P value of enrichment
Data availability statement
The data related to this research can be obtained from the corresponding author upon reasonable request.