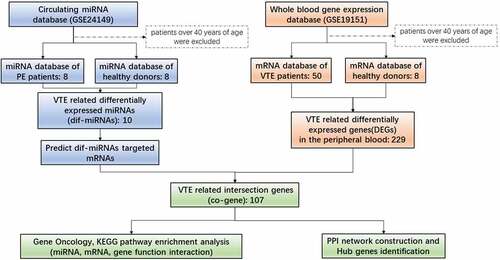
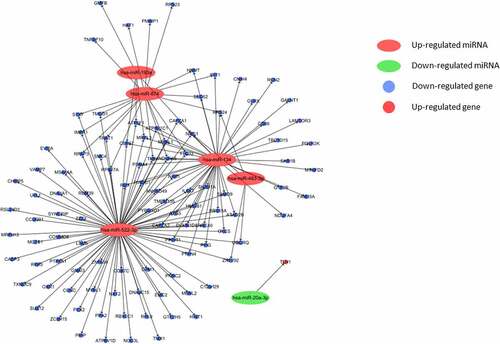
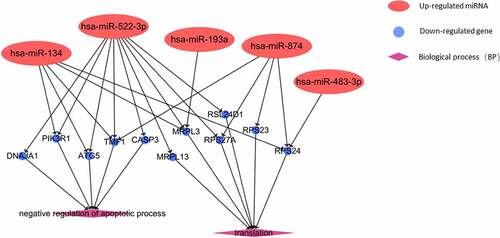
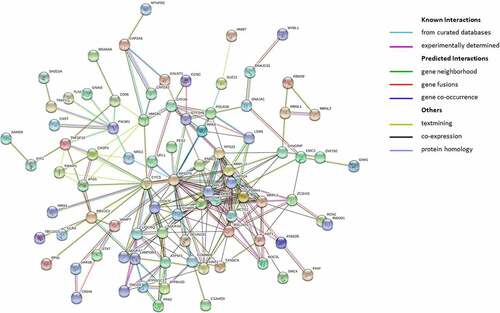
Figures & data
Table 1. Top ten miRNAs with the most significant differential expressions in peripheral blood of VTE patients
Table 2. Differentially expressed genes (DEGs) in peripheral blood of VTE patients
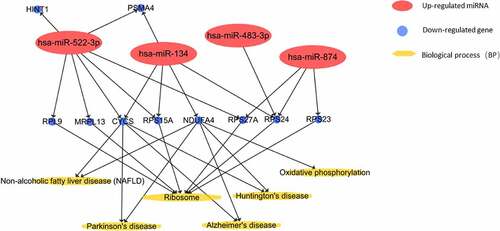
Table 3. Overlapping gene list (co-genes) of dif-miRNAs target genes and DEGs in VTE patients
Table 4. Gene ontology (GO) function enrichment analysis of co-genes in VTE patients
Table 5. KEGG enrichment analysis of in VTE patients
Table 6. Top ten co-genes genes (hub genes) with highest connectivities