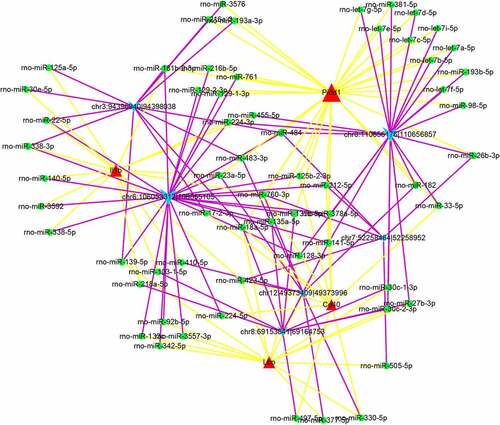
Figures & data
Table 1. The primer sequences in this study
Figure 1. Fluoxetine treatment caused increased weight and reduced cerebral infarction area in rats after MCAO (middle cerebral artery occlusion). (a) The weights of rats in each group were monitored and measured after the experiment. (b) The cerebral infarction area was determined using triphenyltetrazolium chloride staining on the three groups of rats. Sham: the sham group (n = 7), MCAO: the MCAO model group (n = 7), Fluoxetine: the fluoxetine treatment group (n = 7); ** P< 0.01, **** P< 0.0001
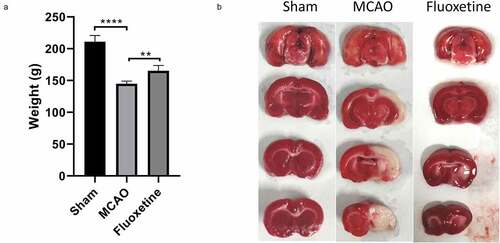
Figure 2. The expression profiles of fluoxetine-mediated circRNAs in the rats’ brain tissue after MCAO (middle cerebral artery occlusion). (a) The back-splicing reads in the sequences of sham, MCAO model, and fluoxetine treatment groups. (b) The length distribution of circRNAs. (c) The genomic origin of circRNAs. The differentially expressed circRNAs were exhibited in heat and volcano maps between the sham and MCAO groups (d and f). The differentially expressed circRNAs were exhibited in heat and volcano maps between the MCAO and fluoxetine treatment groups (e and g)
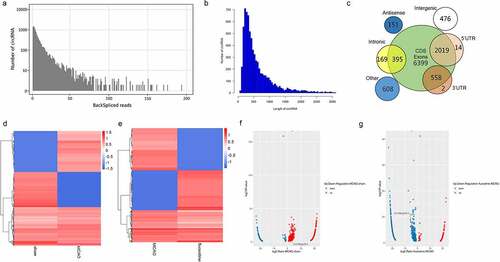
Figure 3. The expression profiles of fluoxetine-mediated mRNAs and functional analysis in the rats’ brain tissue after MCAO (middle cerebral artery occlusion). (a) and (c) is the heat and volcano maps of differentially expressed mRNAs between the sham and MCAO group. (b) and (d) is the heat and volcano maps of differentially expressed mRNAs between the MCAO and fluoxetine treatment groups. (e) KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis was used to show the top 20 signaling pathways between the sham and MCAO model groups. (f) KEGG analysis was used to show the top 20 signaling pathways between the MCAO and fluoxetine treatment groups
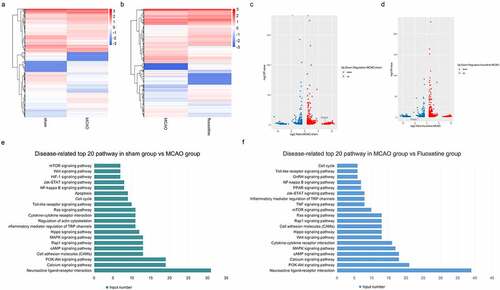
Figure 5. The identification of the candidate circRNAs and mRNAs. (a) qRT-PCR (quantitative reverse transcriptase polymerase chain reaction) detected the expression of circHipk3, circMap2k1, circSipa1l1, and circNav3 in the sham, MCAO model, and fluoxetine treatment groups. (b) and (c) Sanger sequencing and agarose gel electrophoresis was used to verify the circular structure of circHipk3, circMap2k1, circSipa1l1, and circNav3. (d) qRT-PCR detected the four candidate mRNAs, Cd40 (CD40 molecule), Pidd1 (P53-induced death domain protein 1), Il1b (interleukin 1 beta), and LBP (lipopolysaccharide binding protein), that had competitive RNA effect with circRNAs circHipk3, circMap2k1, circSipa1l1, and circNav3. * P< 0.05, ** P< 0.01, ** P< 0.001, **** P< 0.0001
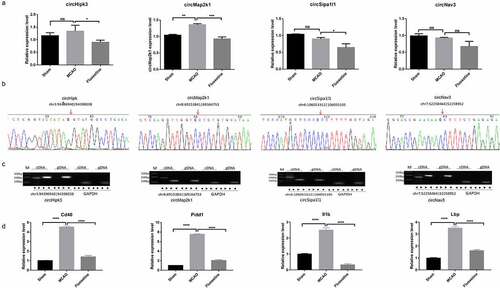
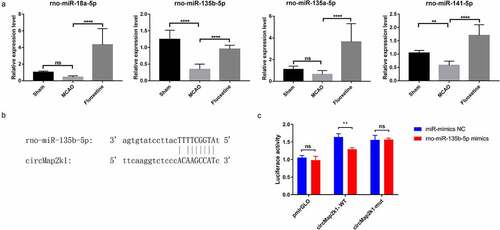
Figure 6. The effect of fluoxetine on circMap2k1/miR-135b-5p/Pidd1 axis. (a) Quantitative reverse transcriptase polymerase chain reaction detected miRNAs rno-miR-18a-5p, rno-miR-135b-5p, rno-miR-135a-5p, and rno-miR-141-5p that has binding sites with circMap2k1 and Pidd1 (P53-induced death domain protein 1). (b) TargetScan predicted the binding sites of rno-miR-135b-5p and circMap2k1. (c) Dual-luciferase reporter assay verified the binding of rno-miR-135b-5p and circMap2k1. ** P< 0.01, **** P< 0.0001
Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.