Figures & data
Figure 1. The prediction of pivotal miRNAs in VSMCs which participated in the pathogenesis of AS
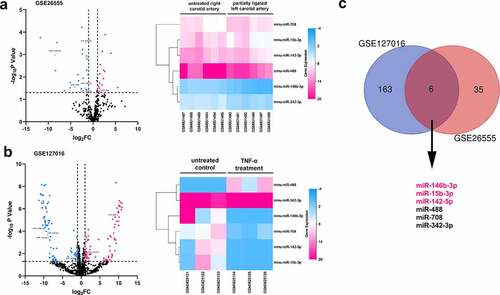
Figure 2. The effect of PDGF treatment on the expression of miR-146b-3p in VSMCs
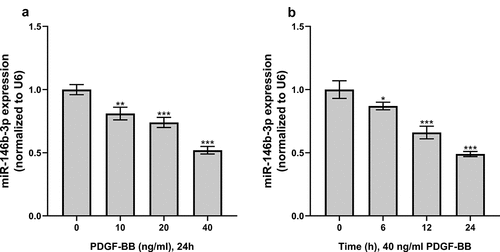
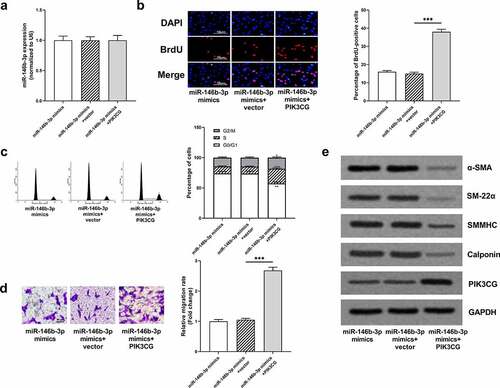
Figure 3. The effect of miR-146b-3p on VSMCs
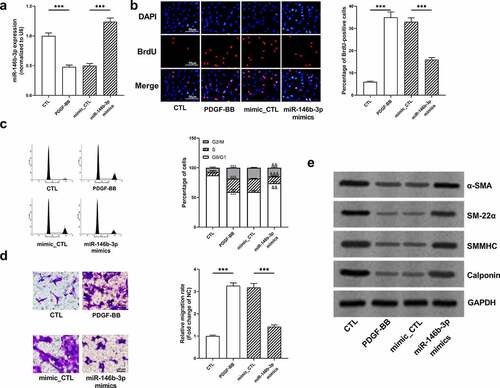
Figure 4. The prediction of pivotal genes in VSMCs which participated in the pathogenesis of AS
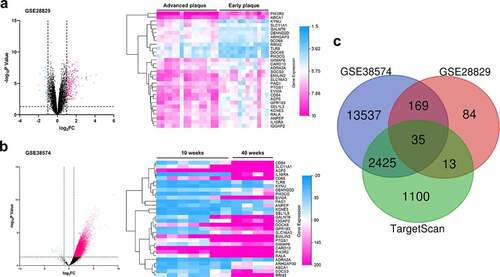
Figure 5. The expression of PIK3CG was inhibited by miR-146b-3p in VSMCs
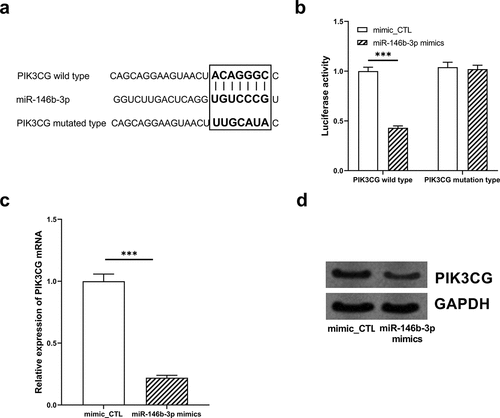
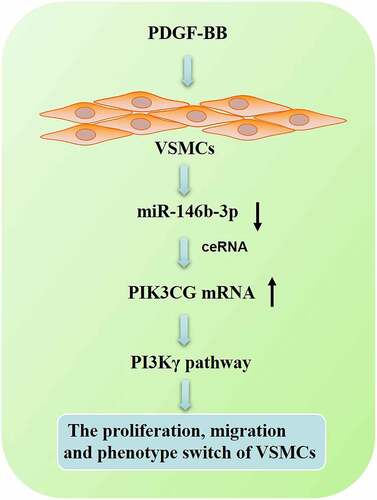
Figure 6. The effects of miR-146b-3p on VSMCs were abrogated by PIK3CG