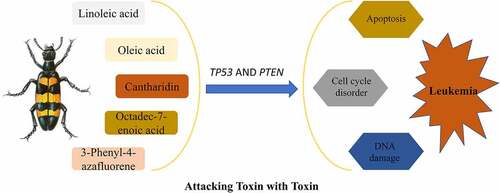
Figures & data
Table 1. The effective components of Mylabris screened using TCMSP database
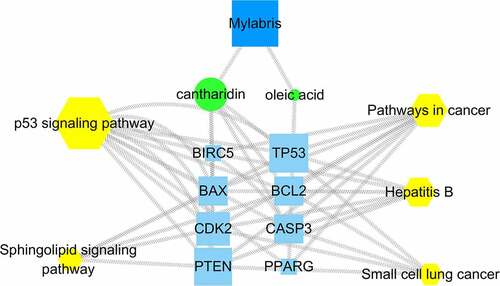
Figure 1. Interaction diagram between active ingredients and potential targets
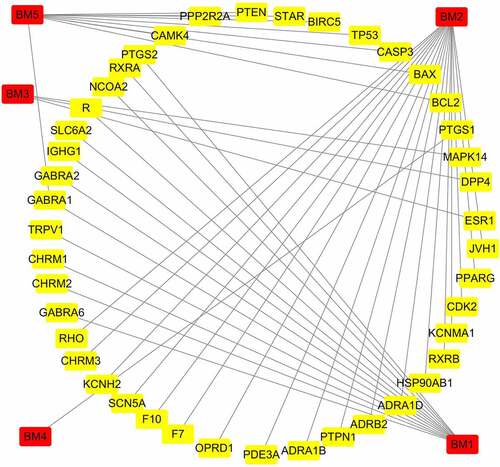
Figure 2. Intersection of mylabris target genes and leukemia disease genes
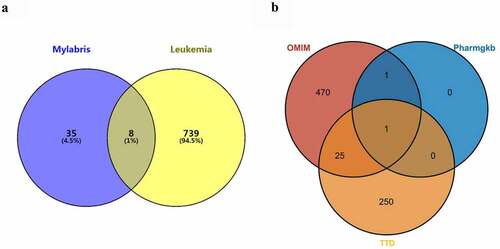
Figure 3. PPI network interaction among intersection targets
Figure 4. GO enrichment analysis of 8 potential targets
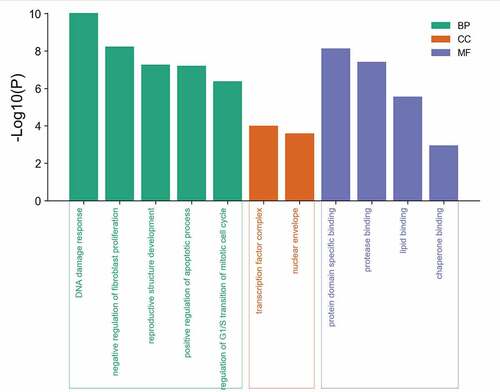
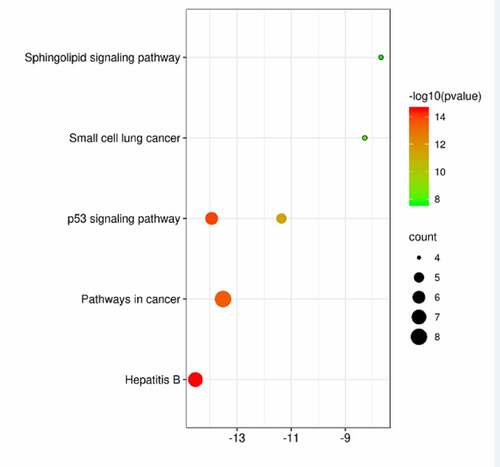
Figure 5. KEGG pathway enrichment analysis of potential acting genes
Data availability statement
All of the datasets analyzed were acquired from the traditional Chinese medicine system pharmacology (TCMSP) database (https://tcmspw.com/tcmsp.php), Online Mendelian Inheritance in Man (OMIM) database (http://www.omim.org/), Therapeutic Target Database (TTD) database (http://www.db.idrblab.net/ttd/), PharmGKB database (https://www.pharmgkb.org/) and GeneCards database (https://www.genecards.org).