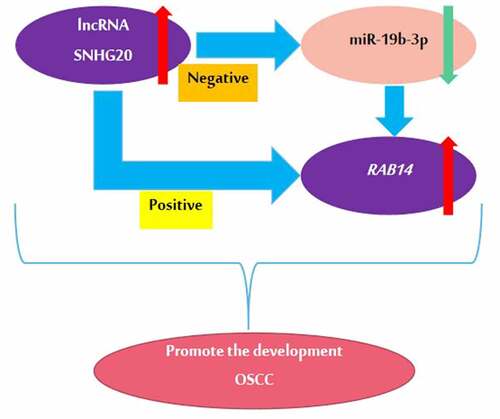
Figures & data
Figure 1. LncRNA SNHG20 was overexpressed in OSCC tissues and CAL27 and SCC25 cells
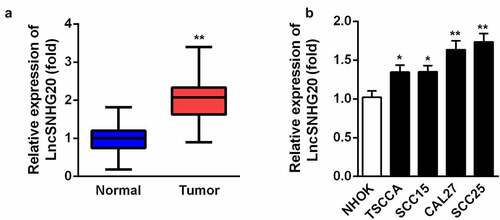
Figure 2. The knockdown of lncRNA SNHG20 decreased the proliferation, migration, and invasion of SCC25 and CAL27 cells
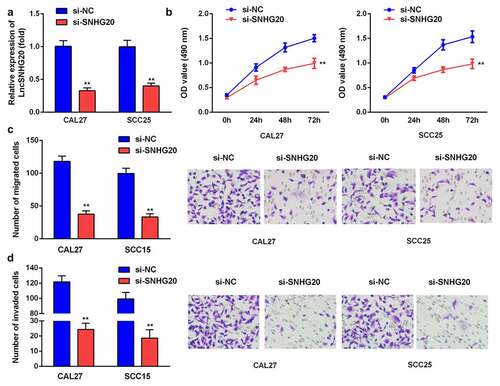
Figure 3. LncRNA SNHG20 regulates the OSCC cell functions by sponging miR-19b-3p
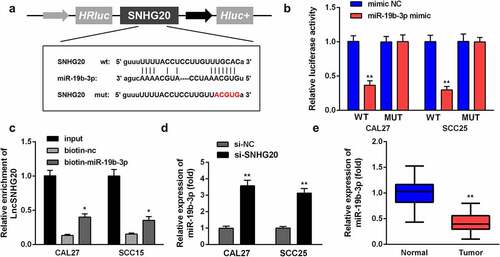
Figure 4. The knockdown of miR-19b-3p reversed the effect of si-SNHG20 on the proliferation, migration, and invasion of SCC25 and CAL27 cells
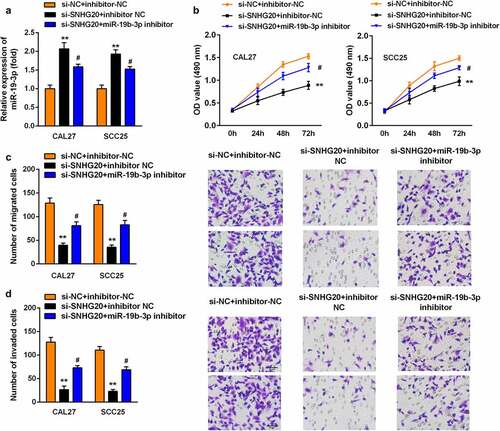
Figure 5. RAB14 is the target gene of miR-19b-3p in OSCC
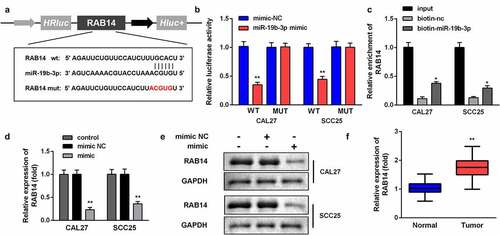
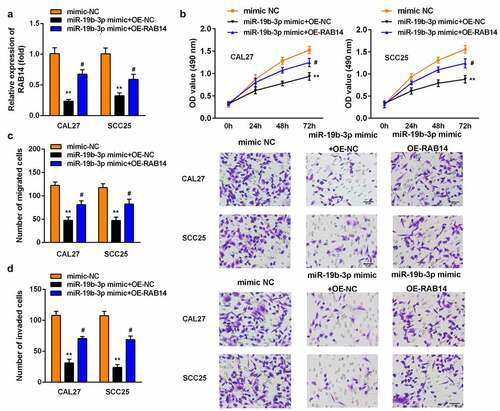
Figure 6. Overexpression of RAB14 reversed the effect of miR-19b-3p mimic on the proliferation, migration, and invasion of SCC25 and CAL27 cells
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.