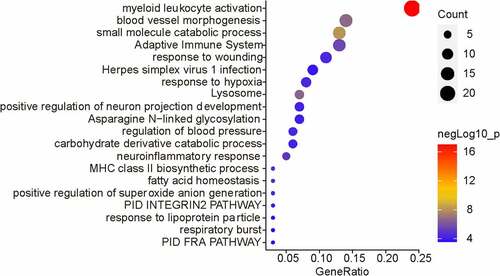
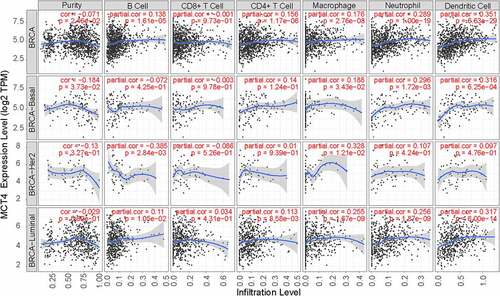
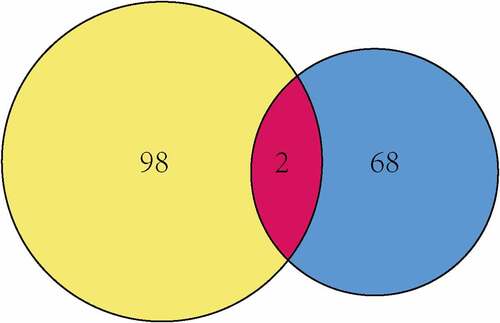
Figures & data
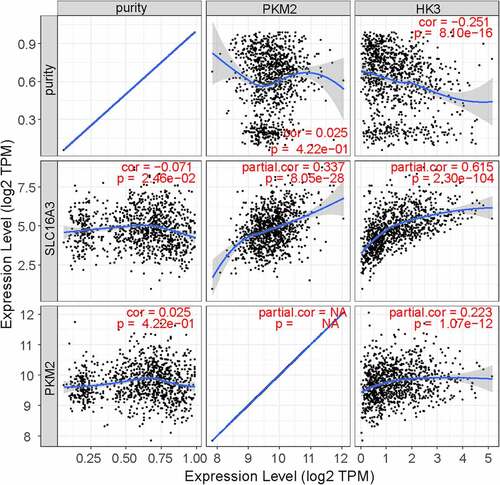
Figure 1. The expression profiles of MCT1 and MCT4 in tumor and normal tissues
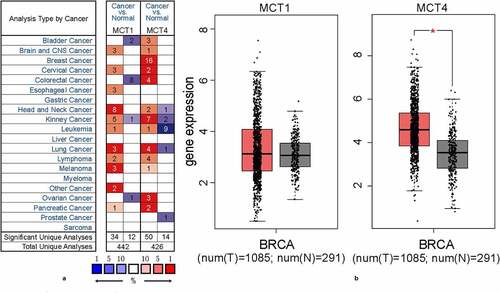
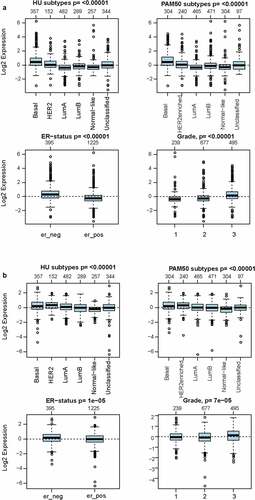
Figure 2. The expression pattern of MCT4 in different subtypes of BC using the GOBO dataset (n = 1881)
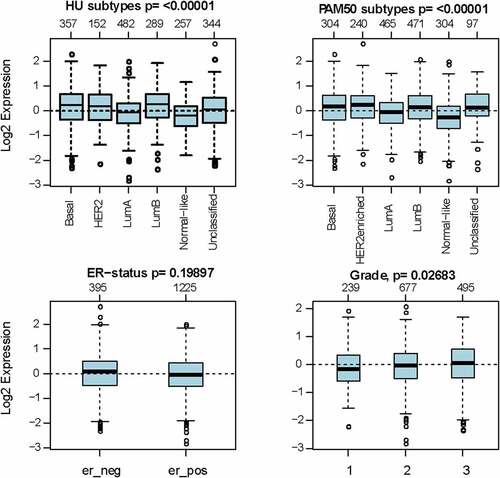
Figure 3. The expression profile of MCT4 in different subtypes of BC using the merged microarray datasets (n = 10,001)
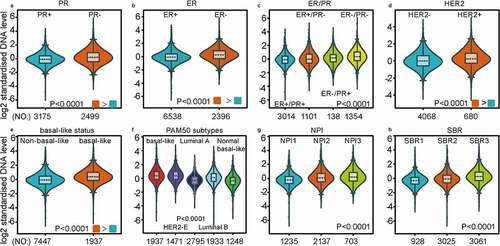
Figure 4. MCT4 expression signature from RNA-seq level exploiting merged RNA-seq datasets (n = 4712)
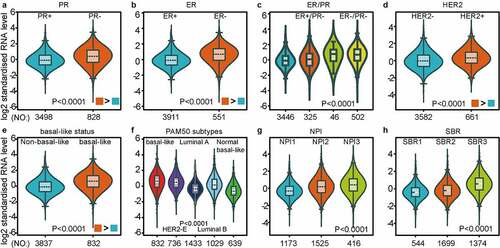
Figure 5. The kaplan-meier survival curves of high and low MCT4 expression in BC through the kaplan-meier plotter database. the RFS in (a) PR+ BC, (b) PR- BC, (c) ER+ BC, (d) ER- BC, (e) HER2+ BC, (f) HER2- BC, (g) basal-like subtype, (h) luminal A subtype, (i) luminal B subtype. RFS: relapse-free survival; HR: hazard ratio
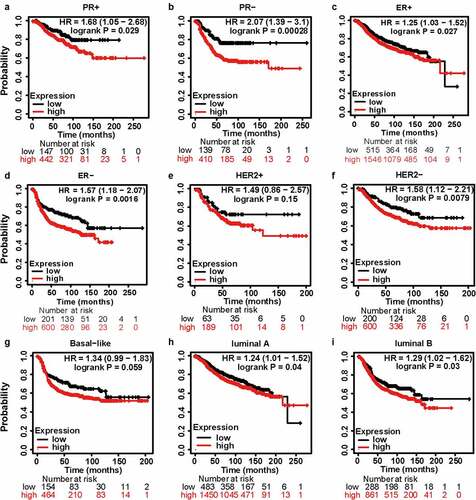
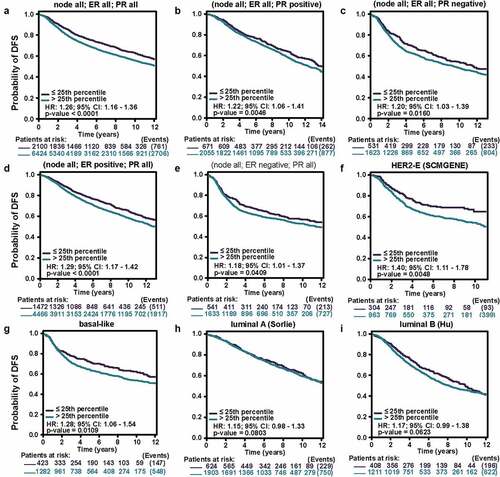
Figure 6. The kaplan-meier survival curves of high and low MCT4 expression in BC through the bc-GenExMiner v4.5