Figures & data
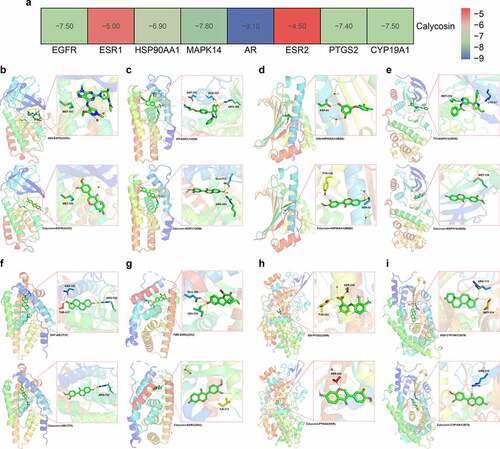
Figure 1. (a) As showed in Venn diagram assay, all candidate, mapped targets of calycosin and BPA-related OS were identified and produce network map using shared targets. (b) After further bioinformatics analysis, all 9 core targets of calycosin in the treatment of BPA-related OS were identified accordingly
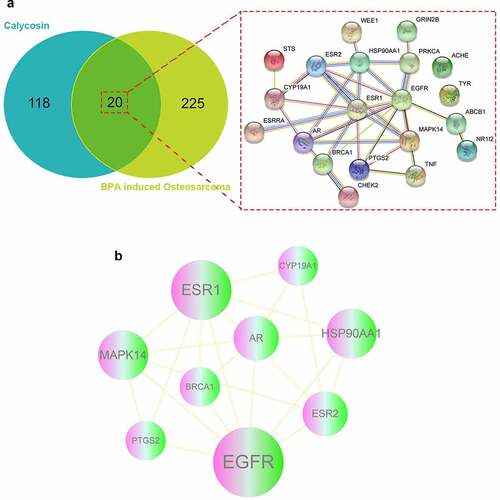
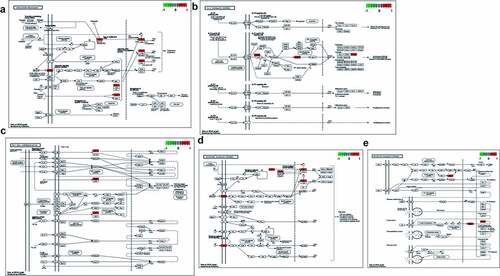
Figure 2. (a) After enrichment analyses, top GO-based functional processes, including bone, immunity, inflammation, hormone and cancer, were revealed. (b) Other top KEGG-based molecular mechanisms of calycosin to treat BPA-related OS, including immunity, hormone, cancer and signaling, were uncovered
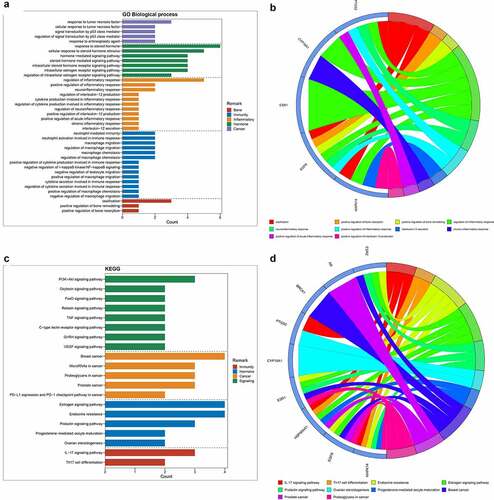
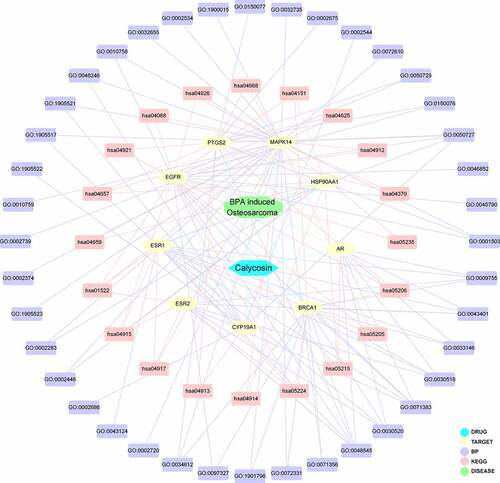
Figure 3. By using integrated analysis, network visualization of calycosin-target-BP-KEGG-BPA/OS was produced and highlighted