Figures & data
Figure 1. Dexmedetomidine improved OGD/R-induced neurological injury. (a) The cell viability of SH-SY5Y cells was tested through CCK-8 assay in the Control, OGD/R-6 h, OGD/R-12 h, OGD/R-24 h, OGD/R-48 h group. (b) SNHG11 expression was examined through RT-qPCR in the Control, OGD/R-6 h, OGD/R-12 h, OGD/R-24 h, OGD/R-48 h group. (c) The cell viability was measured through CCK-8 assay after treating with Dex (0, 50, 100, 200, and 400 ng/ml). (d) SNHG11 expression was assessed through RT-qPCR after in the Control, OGD/R-6 h, OGD/R-12 h, OGD/R-24 h, OGD/R-48 h group. (e) The cell apoptosis was measured through TUNEL assay in the Control, OGD/R, OGD/R+ saline, and OGD/R+ Dex groups. (f) The levels of SOD, CAT, GSH-Px, and MDA were verified through the appropriate commercial kits in the Control, OGD/R, OGD/R+ saline, and OGD/R+ Dex groups. *P < 0.05
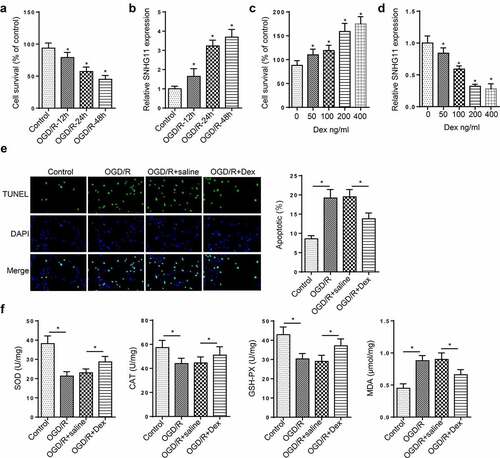
Figure 2. Dexmedetomidine reversed the effects of SNHG11 on OGD/R-induced neurological injury. Groups were divided into the OGD/R+ oe-NC, OGD/R+ oe-SNHG11, and OGD/R+ oe-SNHG11+ Dex groups. (a) The cell viability was measured through CCK-8 assay. (b) The cell apoptosis was detected through TUNEL assay. (c) The levels of SOD, CAT, GSH-PX, and MDA were confirmed through the appropriate commercial kits. *P < 0.05
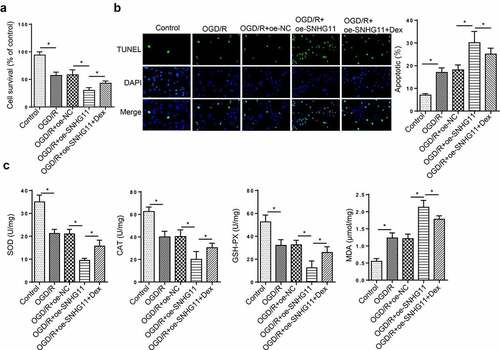
Figure 3. miR-324-3p is a target of SNHG11. (a) The potential miRNAs that could bind with SNHG11 were predicted through starBase website with the condition (CLIP Data: high stringency (≥3)). The binding ability of miRNAs to SNHG11 was confirmed through RNA pull-down assay. (b) MiR-324-3p expression was tested through RT-qPCR. (c) The overexpression efficiency of miR-324-3p was verified through RT-qPCR. (d) The binding ability between SNHG11 and miR-324-3p was detected through the luciferase reporter assay. *P < 0.05
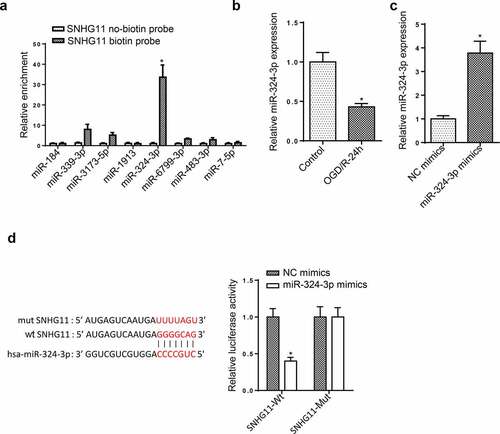
Figure 4. SNHG11 regulated VEGFA level by targeting miR-324-3p. (a) The downstream target genes (n = 11) were selected through starBase website with the condition (Degradome Data: high stringency (≥3)). The levels of target genes were confirmed after miR-324-3p mimics through RT-qPCR. (b) VEGFA expression was measured through RT-qPCR. (c) The binding ability between miR-324-3p and VEGFA was verified through the luciferase reporter assay. (d) The enrichment of SNHG11, miR-324-3p, and VEGFA was evaluated in RISC complex through RIP assay. *P < 0.05
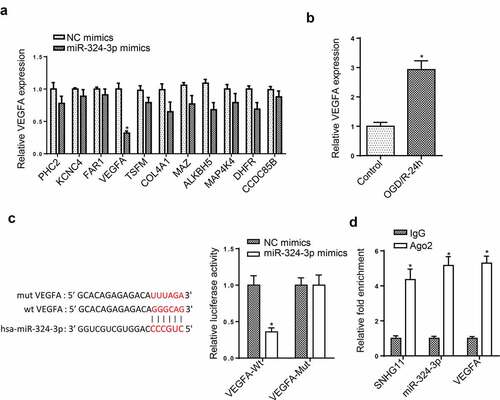
Figure 5. SNHG11 affected OGD/R-induced neurological injury by regulating VEGFA. Groups were divided into the OGD/R+ sh-NC, OGD/R+ sh-SNHG11, and OGD/R+ sh-SNHG11+ oe-VEGFA groups. (a) The cell viability was measured through CCK-8 assay. (b) The cell apoptosis was confirmed through TUNEL assay. (c) The levels of SOD, CAT, GSH-PX, and MDA were confirmed through the appropriate commercial kits. *P < 0.05
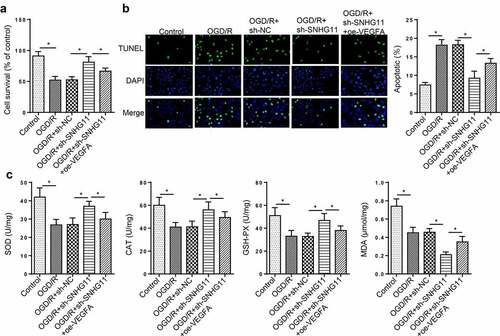
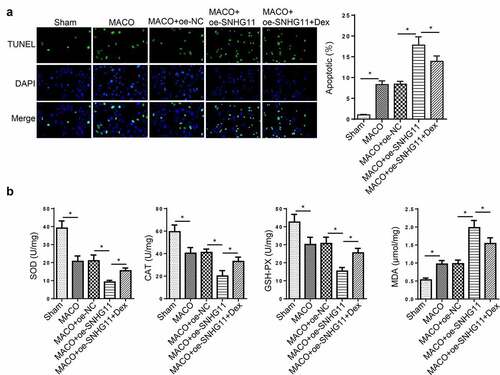
Figure 6. Dexmedetomidine improved neurological injury by regulating SNHG11 in vivo. Groups were divided into the Sham, MCAO, MCAO +oe-SNHG11 and MCAO +oe-SNHG11+ Dex group. (a) The cell apoptosis was examined through TUNEL assay in the hippocampus of rat model. (b) The levels of SOD, CAT, GSH-PX, and MDA were confirmed through the appropriate commercial kits in the hippocampus of rat model. *P < 0.05