Figures & data
Table 1. Primers for osteogenesis differentiation markers and circRNAs
Table 2. siRNAs’ sequences
Figure 1. Induction of BMSCs osteogenic differentiation. (a) BMSCs’ cell morphology switched from elongated fusiform to polygonal (Magnification: 200×, scale bar: 250 μm). (b) Alizarin Red staining showed increased mineralized nodules formation (Magnification: 200×, scale bar: 250 μm). (c) ALP staining showed increased ALP protein expression (Magnification: 200×, scale bar: 250 μm). (d) ALP activity tests showed increased ALP activity (***p < 0.001). (e) qRT-PCR showed increased the mRNA expression of RUNX2, OPN, and OCN (**p < 0.01, ****p < 0.0001). Non-ind: Non-induced group; Ind: Induced group
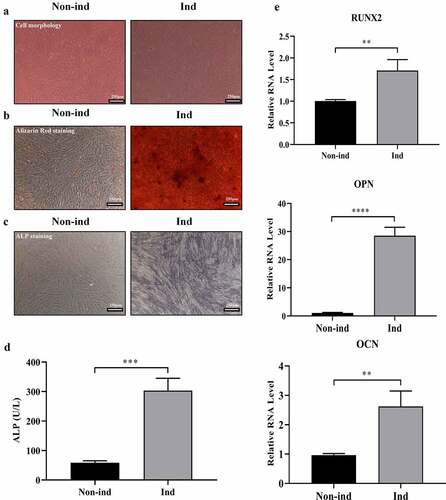
Figure 2. (a) Differentially expressed cirRNAs profile and function analysis. Venn diagram showed the number of circRNAs detected in Non-ind group and Ind group. (b) Chromosomes distribution of circRNAs. Non-ind group: inner green circle; Ind group: outer red circle. (c) Heat map of differential circRNAs in Non-ind group and Ind group. (d) GO analysis result of the parent gene of differentially expressed circRNAs in biological process, cellular component, molecular function. (e) KEGG analysis result of the parent gene of differentially expressed circRNAs. (f) GSEA analysis result of the parent genes of differentially expressed circRNAs being involved in osteogenic differentiation-related pathways. (g) Heat map of the parent genes of differentially expressed circRNAs in GSEA analysis
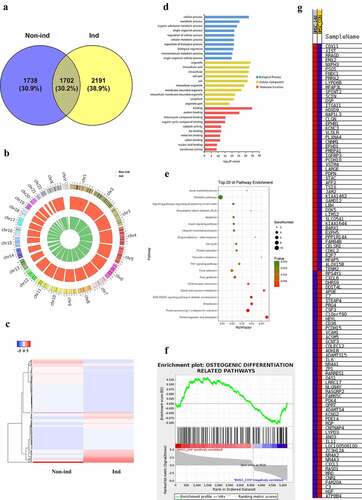
Figure 3. The validation of most differentially expressed circRNAs and construction of ceRNA networks (a). Circ_0109177, circ_0001085, circ_0005078, circ_0002075, circ_0000471, circ_0006083, circ_0006482, circ_0000707, circ_0005564, and circ_0005255 were determined by qRT-PCR. All experiments were performed in triplicate (*p < 0.05). (b) The ceRNA networks of five differentially expressed circRNAs with more than one binding miRNA site
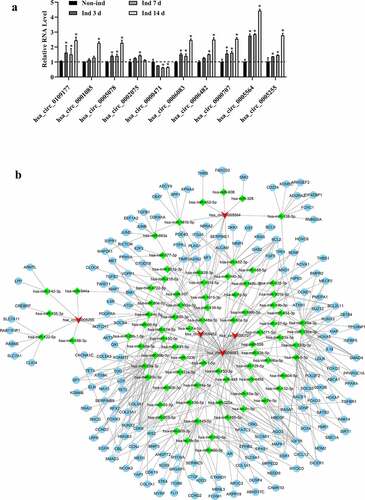
Figure 4. Effects of circ_0005564 on the expression of osteogenic markers. (a) siRNAs was transfected into BMSCs, and circRNAs expression was tested by qRT-PCR. *p < 0.05. (b) RUNX2, OPN, and OCN were detected by qRT-PCR after circ_0005564 silencing. *p < 0.05. (c) The formation pattern of circ_0005564. (d) Divergent and convergent primers were designed to determine the circular form of circ_0005564. PCR products were observed through agarose gel electrophoresis. (e) Sanger sequencing of products from divergent primers of circ_0005564
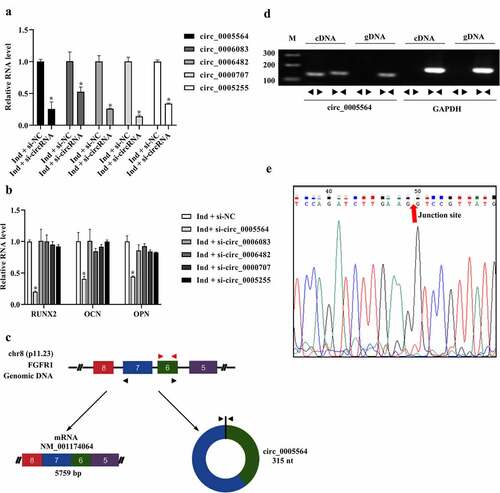
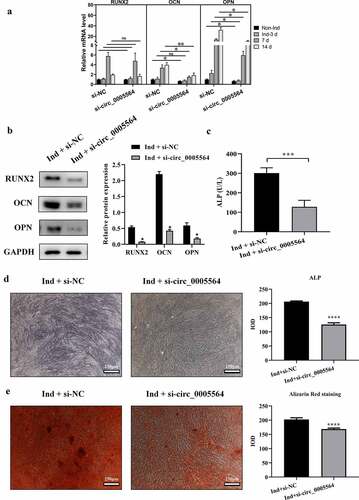
Figure 5. Circ_0005564 silencing inhibited BMSCs osteoblast differentiation. (a) qRT-PCR results of RUNX2, OCN, and OPN mRNA expression after circ_0005564 knockdown in BMSCs after induction at indicated time-points. *p < 0.05, **p < 0.01, N = 3. (b) The protein expression of RUNX2, OPN, and OCN were significantly decreased by circ_0005564 knockdown (*p < 0.05); (c) knockdown circ_0005564 significantly reduced ALP activity in BMSCs with osteogenic differentiation induction (***p < 0.001). (d) circ_0005564 knockdown reduced ALP protein expression in BMSCs with osteogenic differentiation induction. (e) alizarin red staining showed that circ_0005564 knockdown inhibited mineralized nodules formation in BMSCs with osteogenic differentiation induction