Figures & data
Table 1. The information of primers
Figure 1. Characterization of circNfix in cardiomyocytes. (a) CircNfix was back-spliced by exons 2 of Nfix gene as the black arrow showed. The red arrow indicates head-to-tail splicing junction site of circNfix. (b) The expressions of linear mRNA Nfix and circNfix in RNase R-treated cardiomyocytes were detected with RT-qPCR. (c) RT-qPCR analysis of circNfix and Nfix mRNA in actinomycin D-treated cardiomyocytes. (d) CircNfix has been validated using nucleic acid electrophoresis. (e) The distribution of circNfix in cardiomyocytes was examined by RT-qPCR. (f, g) RT-qPCR was used to detect the expression of circNfix in TAC-treated mice and in Ang II treated cardiomyocytes. **P < 0.01, compared with control group; n = 3
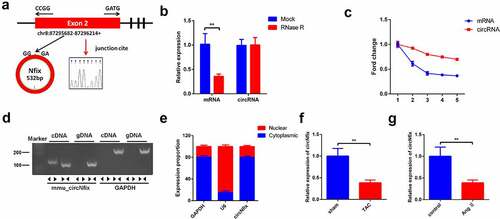
Figure 2. CircNfix protected cardiomyocytes against Ang II in vitro. (a) Transduction efficiency of adenovirus in vitro has been examined by RT-qPCR. (b) The immunofluorence staining was used to analyze the size of cardiomyocytes in different group. **P < 0.01; n = 3
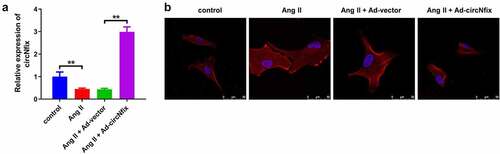
Figure 3. CircNfix protected cardiomyocytes against Ang II in vitro via downregulation of ANP, BNP and β-MHC. (A, B and C) The expressions of cardiac hypertrophic markers including ANP, BNP and β-MHC were evaluated by RT-qPCR. (D, E, F and G) The expressions of cardiac hypertrophic markers including ANP, BNP and β-MHC were detected with Western blotting. **P < 0.01; n = 3
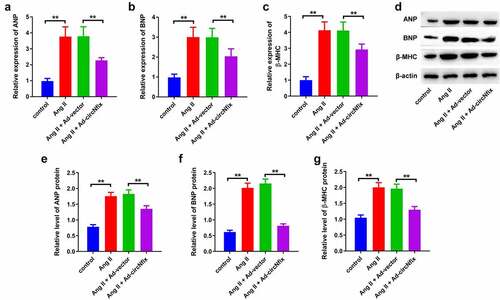
Figure 4. CircNfix protected mice against TAC-induced cardiac hypertrophy. (a) The expression of circNfix in myocardium was analyzed by RT-qPCR. (b) HE staining was performed to evaluate the size of the heart and cardiomyocytes. **P < 0.01; n = 3
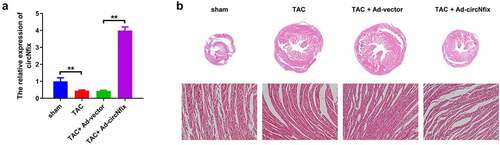
Figure 5. CircNfix protected mice against TAC-induced cardiac hypertrophy via alleviation of cardiac function. (a) Quantification of HW/TL was calculated. (b, c) Examination of the echocardiographic parameters was performed. (d) Cell apoptosis was calculated using TUNEL assay. **P < 0.01; n = 3
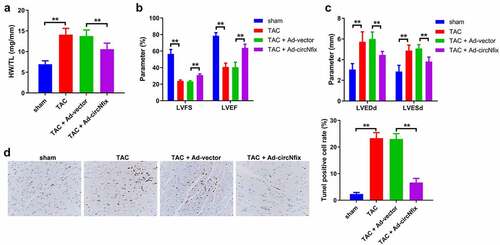
Figure 6. CircNfix sponged miR-145-5p in cardiomyocytes. (a) The predicted binding sequence between circNfix and miR-145-5p. (b) Luciferase assay was performed to detect the interaction between circNfix and miR-145-5p. (c) RNA pull down was performed to detect the interaction between circNfix and miR-145-5p. (d) FISH assay with specific probes targeting miR-145-5p and circNfix has been conducted to detect the co-location of miR-145-5p and circNfix in cardiomyocytes. **P < 0.01; n = 3
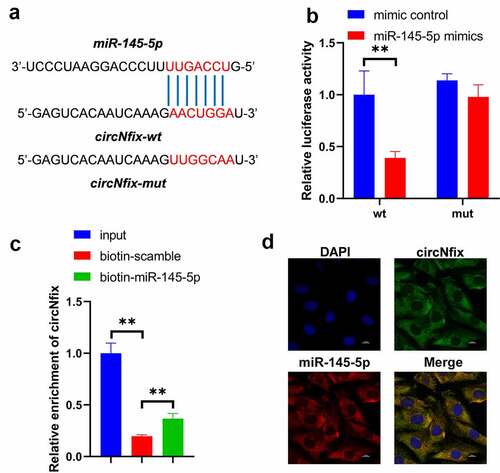
Figure 7. MiR-145-5p mimics could reverse the protective effects of circNfix in Ang II-treated cardiomyocytes. (a) The expressions of cardiac hypertrophic markers including ANP, BNP and β-MHC were evaluated by RT-qPCR. (b) The immunofluorence staining was used to analyze the size of cardiomyocytes in different group. **P < 0.01; n = 3
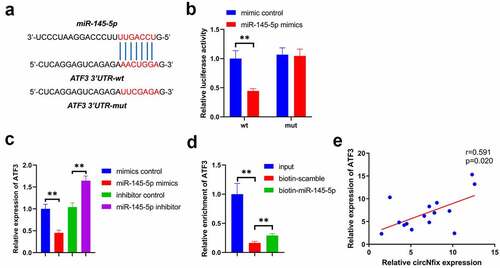
Figure 8. ATF3 was a target of miR-145-5p. (a) The predicted binding sequence between miR-145-5p and ATF3 was presented. (b) Luciferase assay was performed to detect the interaction between miR-145-5p and ATF3. (c) RNA pull down was performed to detect the interaction between miR-145-5p and ATF3. (d) Pearson’s correlation analysis was performed to detect the correlation between ATF3 and circNfix. **P < 0.01; n = 3
Data availability
All the data are available from the corresponding author due to reasonable request.