Figures & data
Table 1. Correlations between RECQL4 level and clinicopathological characteristics
Figure 1. RECQL4 expression in OXA-resistant COAD. (a and b) The UALCAN and GEPIA websites were used to analyze RECQL4 expression between COAD and para-cancer tissues. (c) RECQL4 expression in various types of cancer tissues and corresponding normal tissues, as provided by GEPIA website. (d) RECQL4 mRNA levels in 60 pairs of normal tissues and COAD tissues (OXA-sensitive COAD tissues: n = 36; and OXA-resistant COAD tissues: n = 24) were measured by RT-qPCR. (e) RECQL4 protein expression in OXA-sensitive COAD tissues (n = 36) and OXA-resistant COAD tissues (n = 24) were detected by IHC. (f) RECQL4 mRNA levels in normal human colon epithelial cell line (NCM460), as well as COAD cell lines (SW-480, HT-29, T-84, LS-174 T, DLD-1, and LoVo) were assessed by RT-qPCR. (g) OXA IC50 was determined in the established OXA-resistant COAD cells (T-84/OXA and DLD-1/OXA) and parental COAD cells (T-84 and DLD-1) by CCK-8 assay. (h and i) RECQL4 mRNA and protein levels in T-84/OXA and DLD-1/OXA cells, as well as T-84 and DLD-1 cells were detected by RT-qPCR and Western blotting. The results are presented as the mean ± standard deviation (SD) from at least three independent experiments. *P < 0.05
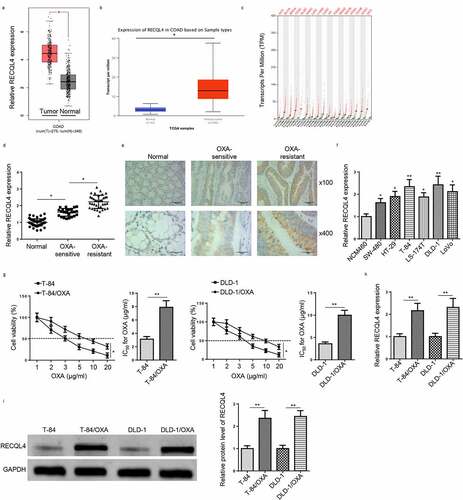
Figure 2. Overexpression of RECQL4 promotes proliferation and metastasis of OXA-resistant COAD cells. (a and b) RECQL4 mRNA and protein levels in T-84/OXA and DLD-1/OXA cells transfected with pcDNA3.1/RECQL4 or pcDNA3.1 were assessed by RT-qPCR and Western blotting. (c) IC50 value of OXA was determined in T-84/OXA and DLD-1/OXA cells treated with pcDNA3.1/RECQL4 or pcDNA3.1 by CCK-8 assay. (d) CCK-8 was used to detect cell viability. (e and f) Flow cytometry was utilized to detect cell apoptosis and cell cycle. (g and h) Wound healing and Transwell assay were utilized to assess cell migration and invasion. The results are presented as the mean ± standard deviation (SD) from at least three independent experiments. *P < 0.05
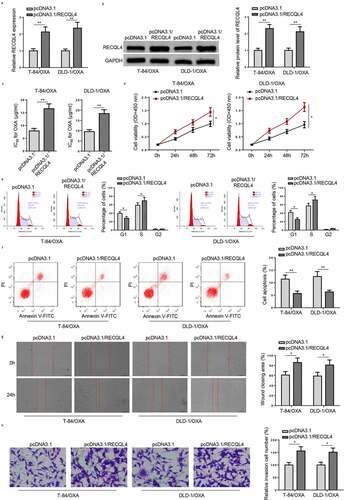
Figure 3. RECQL4 knockdown inhibits OXA-resistant COAD cell proliferation and metastasis. (a and b) RECQL4 mRNA and protein levels in T-84/OXA and DLD-1/OXA cells transfected with sh-RECQL4 or sh-NC were assessed by RT-qPCR and Western blotting. (c) IC50 value of OXA was determined in T-84/OXA and DLD-1/OXA cells treated with sh-NC or sh-RECQL4 by CCK-8 assay. (d) CCK-8 was used to detect cell viability. (e and f) Flow cytometry was utilized to detect cell apoptosis and cell cycle. (g and h) Wound healing and Transwell assay were utilized to assess cell migration and invasion. The results are presented as the mean ± standard deviation (SD) from at least three independent experiments. *P < 0.05
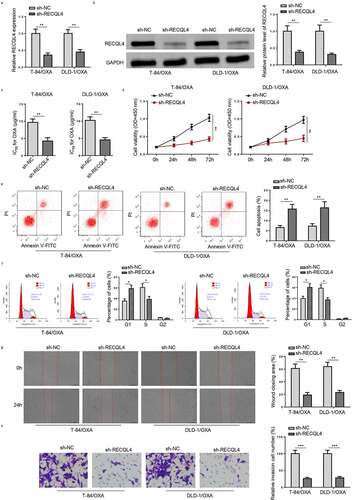
Figure 4. RECQL4 affects the PI3K/AKT signaling in OXA-resistant COAD Cells. (a) Western blotting was used to assess p-PI3K, PI3K, p-AKT, and AKT in T-84/OXA and DLD-1/OXA cells transfected with sh-RECQL4 or sh-NC
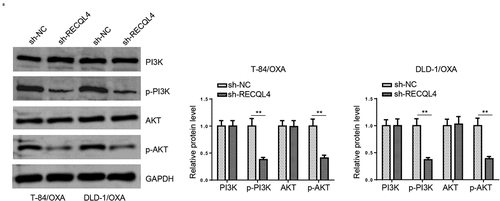
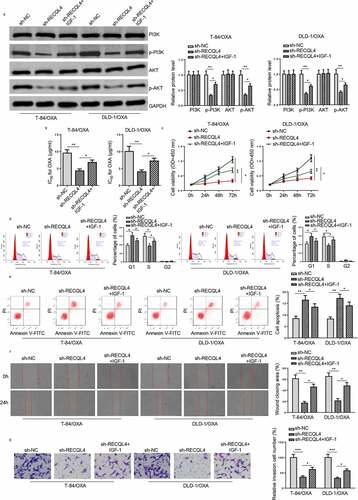
Figure 5. RECQL4 regulates the OXA resistance of COAD cells through the PI3K/AKT signaling. (a) Western blotting was used to assess p-PI3K, PI3K, p-AKT, and AKT in T-84/OXA and DLD-1/OXA cells transfected with sh-NC, sh-RECQL4, or sh-RECQL4+ IGF-1. (b) IC50 value of OXA was determined in T-84/OXA and DLD-1/OXA cells treated with sh-NC, sh-RECQL4 or sh-RECQL4+ IGF-1 by CCK-8 assay. (c) CCK-8 was used to detect cell viability. (d and e) Wound healing and Transwell assay were utilized to assess cell migration and invasion. (f and g) Flow cytometry was utilized to detect cell apoptosis and cell cycle. The results are presented as the mean ± standard deviation (SD) from at least three independent experiments. *P < 0.05