Figures & data
Table 1. RT-qPCR primer sequences
Figure 1. Elevated circSMEK1 expression in NP
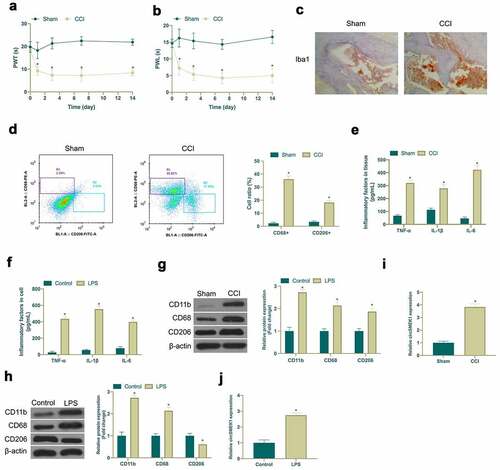
Figure 2. CircSMEK1 knockdown improves NP, while overexpressed circSMEK1 aggravates it
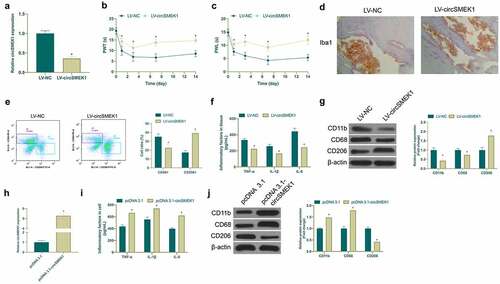
Figure 3. Knocking down miR-216a-5p promotes NP, but overexpressed miR-216a-5p represses it
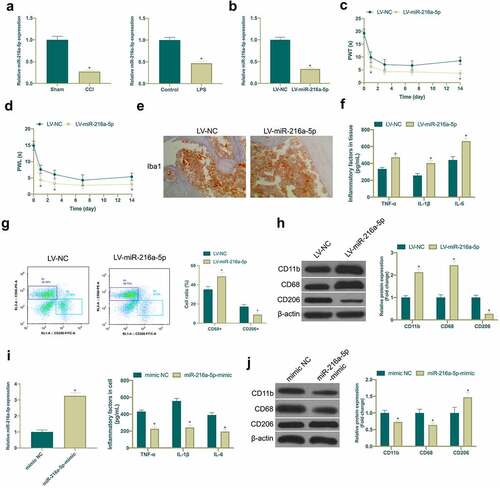
Figure 4. circSMEK1 competitively binds to miR-216a-5p
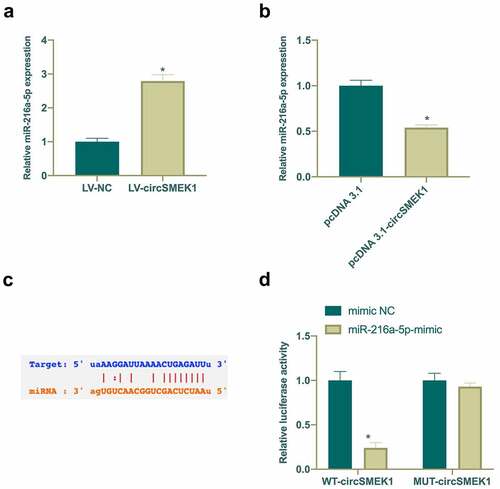
Figure 5. TXNIP is targeted via miR-216a-5p
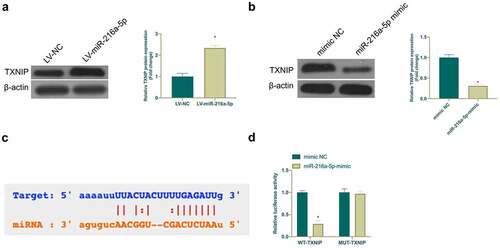
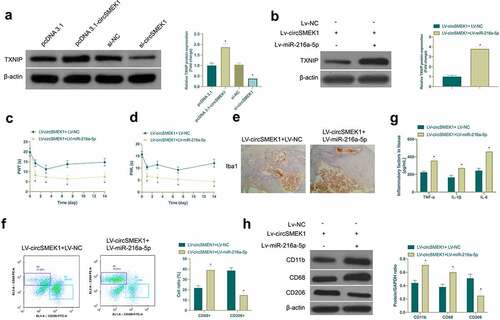
Figure 6. circSMEK1 promotes NP through the miR-216a-5p/TXNIP axis