Figures & data
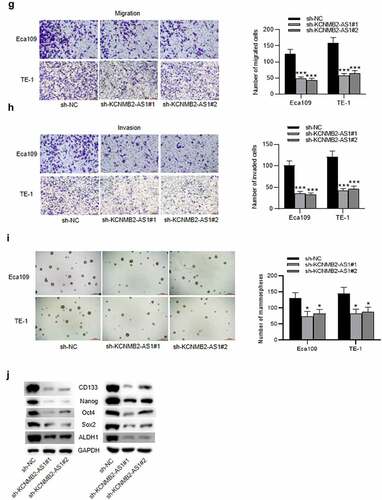
Figure 1. The effects of KCNMB2-AS1 knockdown on ESCA cell growth, motion, and stemness.
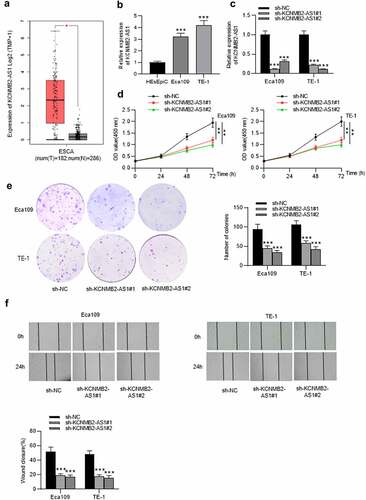
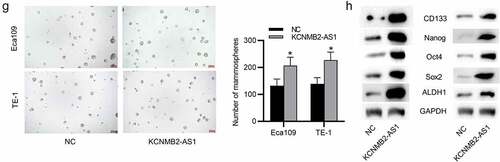
Figure 2. The effects of KCNMB2-AS1 overexpression on ESCA cell growth, motion, and stemness.
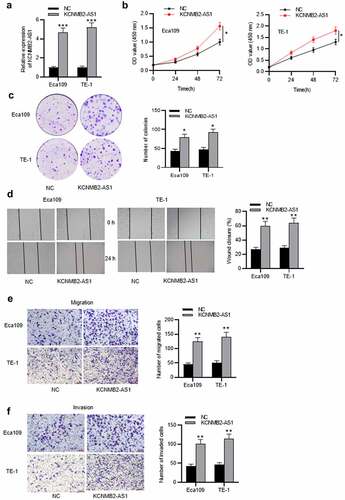
Figure 3. KCNMB2-AS1 binds with miR-3194-3p.
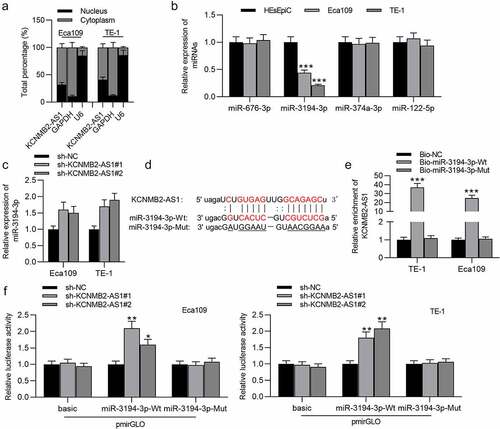
Figure 4. PYGL is targeted by miR-3194-3p.
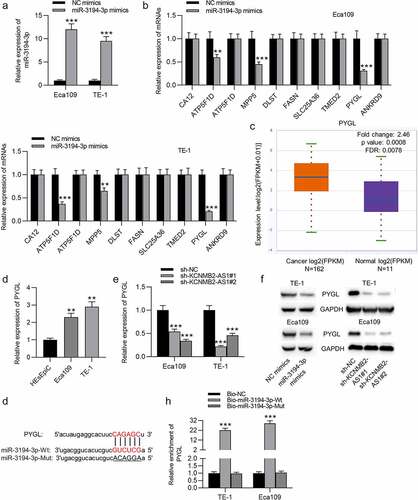
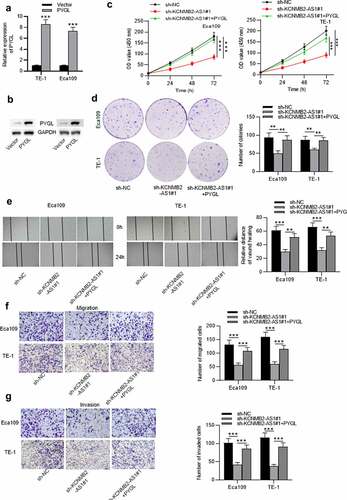
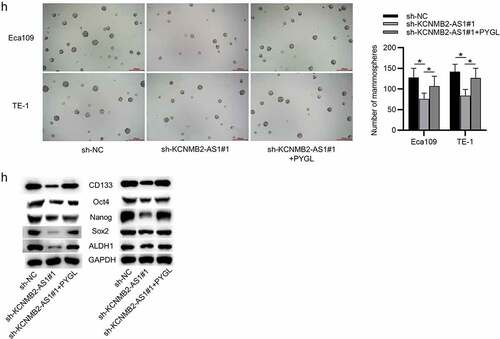
Figure 5. Overexpression of PYGL reverses inhibitory effects of KCNMB2-AS1 knockdown on ESCA cell behavior.